For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHRATATLSAHVPPARSRARTEGRTMPQNQRPYHVAVVGSGPSGFFAAASLLKAADSSDEIEVAVDILEM LPTPWGLVRSGVAPDHPKIKSISKQFEKTAEDPRVRFFGNVVVGEHVTTAELTERYDAVIYAIGAQSDKP LNIPGENLPGSMAAVDFVGWYNAHPNFEHSTPDLSGRRAVVIGNGNVAIDVARILVTDPEVLALTDIADH ALGSLRPRGVEEVVIIGRRGPLQAAFTTLELRELGELEGVDVVVDPAQLEGISDADAEAAGKGVKQNIKV LRGYTERAPRPGHRRIVFKFMTSPVEIKGTDRVEQIVLGHNELVTDEDGWVSAKDTGEREEVPVQLVVRS VGYRGVPTPGLPFDQKCGIIPNTAGRVQGSSNEYVVGWIKRGPTGVIGTNKKDSQDTVDTLMADLTAAEL NTFPSSHTADLADWLSSRQPELVTAEHWKSIDHFERAAGEALGRPRVKLPNVTEMLRIGHA
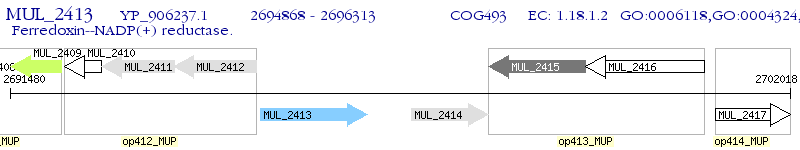
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2413 | fprA_1 | - | 100% (481) | NADPH:adrenodoxin oxidoreductase FprA_1 |
| M. ulcerans Agy99 | MUL_2699 | fprA | e-179 | 73.92% (418) | NADPH:adrenodoxin oxidoreductase FprA |
| M. ulcerans Agy99 | MUL_0264 | fprB | 1e-83 | 39.33% (478) | NADPH:adrenodoxin oxidoreductase FprB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3133 | fprA | 0.0 | 76.16% (453) | NADPH:adrenodoxin oxidoreductase FprA |
| M. gilvum PYR-GCK | Mflv_4452 | - | 0.0 | 70.04% (454) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv3106 | fprA | 0.0 | 75.94% (453) | NADPH:adrenodoxin oxidoreductase FPRA (NADPH-ferredoxin |
| M. leprae Br4923 | MLBr_00666 | fprA | 0.0 | 77.51% (449) | putative NADPH-ferredoxin reductase |
| M. abscessus ATCC 19977 | MAB_3482 | - | 1e-157 | 62.89% (450) | NADPH-ferredoxin reductase FprA |
| M. marinum M | MMAR_1526 | fprA_1 | 0.0 | 98.25% (456) | NADPH:adrenodoxin oxidoreductase FprA_1 |
| M. avium 104 | MAV_4007 | - | 0.0 | 78.81% (453) | NADPH-ferredoxin reductase fpra |
| M. smegmatis MC2 155 | MSMEG_5681 | - | 2e-80 | 39.74% (453) | ferredoxin/ferredoxin--NADP reductase |
| M. thermoresistible (build 8) | TH_2151 | fprA | 0.0 | 73.50% (449) | NADPH:ADRENODOXIN OXIDOREDUCTASE FPRA (NADPH-FERREDOXIN |
| M. vanbaalenii PYR-1 | Mvan_1910 | - | 0.0 | 74.34% (452) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4452|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1910|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2151|M.thermoresistible__bu --------------------------------------------------
MUL_2413|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1526|M.marinum_M --------------------------------------------------
Mb3133|M.bovis_AF2122/97 --------------------------------------------------
Rv3106|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00666|M.leprae_Br4923 --------------------------------------------------
MAV_4007|M.avium_104 --------------------------------------------------
MAB_3482|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5681|M.smegmatis_MC2_155 MPHVITQSCCSDGSCVYACPVNCIHPSPDEPGFATAEMLYIDPDACVDCG
Mflv_4452|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1910|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2151|M.thermoresistible__bu --------------------------------------------------
MUL_2413|M.ulcerans_Agy99 ------------------------MHRATATLSAHVPPARSRARTEGRTM
MMAR_1526|M.marinum_M -------------------------------------------------M
Mb3133|M.bovis_AF2122/97 --------------------------------------------------
Rv3106|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00666|M.leprae_Br4923 --------------------------------------------------
MAV_4007|M.avium_104 --------------------------------------------------
MAB_3482|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5681|M.smegmatis_MC2_155 ACVSACPVGAIAPDTRLEPRQLPFVEINAAFYPKREGKLPPTSKLAPVIE
Mflv_4452|M.gilvum_PYR-GCK MPPLRP---YYVAIVGSGPSGYFAAASLLKSGGADGNRDVCVDMLEMLPT
Mvan_1910|M.vanbaalenii_PYR-1 MPVMRP---YYVAIVGSGPSGFFAAASLLKFGDSPENRDVRVDMLEMLPT
TH_2151|M.thermoresistible__bu ---------LQVAIVGSGPSGFFAAASLLKSADSAEGRDVRVDMLEMLPT
MUL_2413|M.ulcerans_Agy99 PQNQRP---YHVAVVGSGPSGFFAAASLLKAADSSDEIEVAVDILEMLPT
MMAR_1526|M.marinum_M PQNQRP---YHVAVVGSGPSGFFAAASLLKAADSSDEIEVAVDILEMLPT
Mb3133|M.bovis_AF2122/97 ---MRP---YYIAIVGSGPSAFFAAASLLKAADTTEDLDMAVDMLEMLPT
Rv3106|M.tuberculosis_H37Rv ---MRP---YYIAIVGSGPSAFFAAASLLKAADTTEDLDMAVDMLEMLPT
MLBr_00666|M.leprae_Br4923 ---MRPHHIHHIAIVGSGPSGFFAAASVLKAADASDEINVAVDMLEMLPT
MAV_4007|M.avium_104 ---MRP---YHVAIVGSGPSGFFAAASLLKAADGSEHPDIAVDMLEMLPT
MAB_3482|M.abscessus_ATCC_1997 ---MRA---VHVAIVGAGPSGFFAAGSLLKHTD----FDVHVDMLEMLPT
MSMEG_5681|M.smegmatis_MC2_155 APKITRDGPLTVAIVGSGPAAMYAADELLTQRG------VRVNVFEKLPT
:*:**:**:. :** .:*. . : *:::* ***
Mflv_4452|M.gilvum_PYR-GCK PWGLVRSGVAPDHPKIKSISAQFDKTSADPRFRFFGNVVIGEHVHPDELA
Mvan_1910|M.vanbaalenii_PYR-1 PWGLVRSGVAPDHPKIKSISAQFEKTSLDPRFRFFGNVDVGEHVHPSELA
TH_2151|M.thermoresistible__bu PWGLVRSGVAPDHPKIKSISAQFEKVAADPRFRFFGNITVGDHVRPDELA
MUL_2413|M.ulcerans_Agy99 PWGLVRSGVAPDHPKIKSISKQFEKTAEDPRVRFFGNVVVGEHVTTAELT
MMAR_1526|M.marinum_M PWGLVRSGVAPDHPKIKSISKQFEKTAEDPRIRFFGNVVVGEHVTTAELT
Mb3133|M.bovis_AF2122/97 PWGLVRSGVAPDHPKIKSISKQFEKTAEDPRFRFFGNVVVGEHVQPGELS
Rv3106|M.tuberculosis_H37Rv PWGLVRSGVAPDHPKIKSISKQFEKTAEDPRFRFFGNVVVGEHVQPGELS
MLBr_00666|M.leprae_Br4923 PWGLVRSGVAPDHPKIKSISKQFEKTAEDPRFRFFGNVIVGKHIEPAELA
MAV_4007|M.avium_104 PWGLVRSGVAPDHPKIKSISKQFEKTAADPRFRFFGNVSVGEHVQASELA
MAB_3482|M.abscessus_ATCC_1997 PWGLVRSGVAPDHPKIKSVSAVFEKTAAHPRFRFFGNIEVGAKITAEELA
MSMEG_5681|M.smegmatis_MC2_155 PYGLVRAGVAPDHQSTKRVTELFDRMAAQRGFTFYLNVEVGKHLTHADLL
*:****:****** . * :: *:: : . . *: *: :* :: :*
Mflv_4452|M.gilvum_PYR-GCK ERYDAVIYAIGAQSDRALGIPGEDLPGSVPAVDFVGWYNAHPHFVDMAPD
Mvan_1910|M.vanbaalenii_PYR-1 ERYDAVIYAIGAQADRELGIPGEDLPGSLAAVDFVGWYNAHPYFEGMAPD
TH_2151|M.thermoresistible__bu ERYDAVIYAIGTQADRALNIPGEELPGSVAAVDFVGWYNAHPHFENIDPD
MUL_2413|M.ulcerans_Agy99 ERYDAVIYAIGAQSDKPLNIPGENLPGSMAAVDFVGWYNAHPNFEHSTPD
MMAR_1526|M.marinum_M ERYDAVIYAIGAQSDKPLNIPGENLPGSVAAVDFVGWYNAHPNFEHSTPD
Mb3133|M.bovis_AF2122/97 ERYDAVIYAVGAQSDRMLNIPGEDLPGSIAAVDFVGWYNAHPHFEQISPD
Rv3106|M.tuberculosis_H37Rv ERYDAVIYAVGAQSDRMLNIPGEDLPGSIAAVDFVGWYNAHPHFEQVSPD
MLBr_00666|M.leprae_Br4923 ERYDAVIYAVGAQSDRALNIPGEDLPGSIAAVDFVGWYNAHPNFHERSPD
MAV_4007|M.avium_104 ERYDAVIYAVGAQSDRALNIPGEDLPGSIAAVDFVGWYNAHPNFEEATPD
MAB_3482|M.abscessus_ATCC_1997 SRYDAVIYAVGAQSDKPLGIPGDQLSGCIAAVDVVGWYNANPTYQKVSVD
MSMEG_5681|M.smegmatis_MC2_155 GHHHAVVYAVGAPNDRRLDIEGMDLPGTATATEVVAWINGHPDHTDLPVR
::.**:**:*: *: *.* * :*.* .*.:.*.* *.:* .
Mflv_4452|M.gilvum_PYR-GCK VSGGRAVVVGNGNVAIDVARILVSDPDVLAETDIADHALESLHARGVEEV
Mvan_1910|M.vanbaalenii_PYR-1 LSGGRAVVVGNGNVAIDVARILVSDPDVLAETDIADHALRSLHARGVEEV
TH_2151|M.thermoresistible__bu LSGGRAVVIGNGNVALDVARILVTDPEALAKTDIADHALEALRSRGIEEV
MUL_2413|M.ulcerans_Agy99 LSGRRAVVIGNGNVAIDVARILVTDPEVLALTDIADHALGSLRPRGVEEV
MMAR_1526|M.marinum_M LSGRRAVVIGNGNVAIDVARILVTDPEVLALTDIADHALESLRPRGVEEV
Mb3133|M.bovis_AF2122/97 LSGARAVVIGNGNVALDVARILLTDPDVLARTDIADHALESLRPRGIQEV
Rv3106|M.tuberculosis_H37Rv LSGARAVVIGNGNVALDVARILLTDPDVLARTDIADHALESLRPRGIQEV
MLBr_00666|M.leprae_Br4923 LSGSRAVVIGNGNVALDVTRILITDPDVLAFTDIADHALESLRPRGIEEV
MAV_4007|M.avium_104 LSGSRAVVVGNGNVALDVARILVTDPDVLGLTDIADHALESLRPCGVDEV
MAB_3482|M.abscessus_ATCC_1997 LSGERAVVVGNGNVALDVARILGMDPESLHTTDIADRALDSLDHNAIREV
MSMEG_5681|M.smegmatis_MC2_155 LDHERVVIVGNGNVALDVARVLTSDPDALARTDISDHALAALRSSAVQEV
:. *.*::******:**:*:* **: * ***:*:** :* .: **
Mflv_4452|M.gilvum_PYR-GCK LIIGRRGPLQAPFTTLELRELGDLEALGDVDVIVDPGDFADITDEDLEAA
Mvan_1910|M.vanbaalenii_PYR-1 MIIGRRGPLQAPFTTLELRELGDLEALGDVDIVIDPADFADITDDDLEAA
TH_2151|M.thermoresistible__bu VIVGRRGPLQATFTTLELRELGALEGLADVDVVVDPADFADITDEDLEAA
MUL_2413|M.ulcerans_Agy99 VIIGRRGPLQAAFTTLELRELGELEG---VDVVVDPAQLEGISDADAEAA
MMAR_1526|M.marinum_M VIIGRRGPLQAAFTTLELRELGELEG---VDVVVDPAQLEGISDADAEAA
Mb3133|M.bovis_AF2122/97 VIVGRRGPLQAAFTTLELRELADLDG---VDVVIDPAELDGITDEDAAAV
Rv3106|M.tuberculosis_H37Rv VIVGRRGPLQAAFTTLELRELADLDG---VDVVIDPAELDGITDEDAAAV
MLBr_00666|M.leprae_Br4923 VIVGRRGPLQTAFTTLELRELADIEG---VDVLVDPAQLEGISDENAAAA
MAV_4007|M.avium_104 VVIGRRGPLQTAFTTLELRELGELDN---VDVIVDPAQLEGVTDEDVAAA
MAB_3482|M.abscessus_ATCC_1997 VIIGRRGPLQATFTPLELREMGELPG---VDVVVDPEDLADITDEQLAAA
MSMEG_5681|M.smegmatis_MC2_155 VIAARRGPAASAFT---LPELIGLTS--TCDVVLDSADHDLVLRDLAQET
:: .**** :.** * *: : *:::*. : : .
Mflv_4452|M.gilvum_PYR-GCK GKTVRNNIKVLRGYAETTPKGAKRRIVFRFRTSPIEIR----GDGKVESI
Mvan_1910|M.vanbaalenii_PYR-1 GKTVRNNIKVLRGYADKAPKGARRRIVFRFRTSPIEIK----GGDRVQSI
TH_2151|M.thermoresistible__bu GKTVKQNVKVLRGYAEQGVRGAPRRIVFRFRTSPIEIR----GDGRVQSI
MUL_2413|M.ulcerans_Agy99 GKGVKQNIKVLRGYTERAPRPGHRRIVFKFMTSPVEIK----GTDRVEQI
MMAR_1526|M.marinum_M GKGIKQNIKVLRGYTERAPRPGHRRIVFKFMTSPVEIR----GTDRVEQI
Mb3133|M.bovis_AF2122/97 GKVCKQNIKVLRGYADREPRPGHRRMVFRFLTSPIEIK----GKRKVERI
Rv3106|M.tuberculosis_H37Rv GKVCKQNIKVLRGYADREPRPGHRRMVFRFLTSPIEIK----GKRKVERI
MLBr_00666|M.leprae_Br4923 GKTTRQNIKVLRDYTVRTPKPGHRRIVFRFLTSPIEIK----GKGKVERI
MAV_4007|M.avium_104 GKTAKQNISVLRDYAKRSPRPGHRRIVLRFMTSPIEIK----GDGRVESI
MAB_3482|M.abscessus_ATCC_1997 PKPTRTNIETLRKFSERPHTEGHRRVVFRFRTSPIELHSDSAGVGVVESI
MSMEG_5681|M.smegmatis_MC2_155 DPLTRNKLEILAKLGDASAPITRPRIRFAYQLTPRRVL----GENQVTAV
: ::. * *: : : :* .: * * :
Mflv_4452|M.gilvum_PYR-GCK VLGRNELIRGDDGRISAKDTGERDEVAAQLVVRAVGYRGLPTPGLPFDDR
Mvan_1910|M.vanbaalenii_PYR-1 VLGRNELTKGADGRVSAADTGEREEVPAQLVVRAVGYRGLPTPGLPFDER
TH_2151|M.thermoresistible__bu VLGRNELVSDATGRIVAKDTGAREELPAQLVVRAIGYRGLPTPGLPFDER
MUL_2413|M.ulcerans_Agy99 VLGHNELVTDEDGWVSAKDTGEREEVPVQLVVRSVGYRGVPTPGLPFDQK
MMAR_1526|M.marinum_M VLGHNELVTDEDGWVSAKDTGEREELPVQLVVRSVGYRGVPTPGLPFDQK
Mb3133|M.bovis_AF2122/97 VLGRNELVSDGSGRVAAKDTGEREELPAQLVVRSVGYRGVPTPGLPFDDQ
Rv3106|M.tuberculosis_H37Rv VLGRNELVSDGSGRVAAKDTGEREELPAQLVVRSVGYRGVPTPGLPFDDQ
MLBr_00666|M.leprae_Br4923 VLGQNELVTDDNGRVAAKDTGVREELPAQLIVRSIGYRGVPTPGLPFDDS
MAV_4007|M.avium_104 VLGRNELVTDETGRVLARDTGEREELPVQLVVRSVGYRGVPTPGLPFDEK
MAB_3482|M.abscessus_ATCC_1997 TVGRNKLVE-EDGYVVAHDTGERQTLPTHLVVRAIGYRGVPVPGLPFDSR
MSMEG_5681|M.smegmatis_MC2_155 EFG-------------VTGTDEVRTLDAGLLLTSIGYRGKAIADLPFDED
.* . .*. : . *:: ::**** . ..****.
Mflv_4452|M.gilvum_PYR-GCK AGTIPHVDGKVEGRRN------DYVVGWIKRGPTGVIGSNKKDSADTVAT
Mvan_1910|M.vanbaalenii_PYR-1 AGTIPHTDGRVAGSRN------EYVVGWIKRGPTGVIGSNKKDSADTVDT
TH_2151|M.thermoresistible__bu SGTIPHTDGRIEGSRN------EYVVGWIKRGPSGVIGSNKKDSQDTVDT
MUL_2413|M.ulcerans_Agy99 CGIIPNTAGRVQGSSN------EYVVGWIKRGPTGVIGTNKKDSQDTVDT
MMAR_1526|M.marinum_M RGIIPNTAGRVQGSSN------EYVVGWIKRGPTGVIGTNKKDSQDTVDT
Mb3133|M.bovis_AF2122/97 SGTIPNVGGRINGSPN------EYVVGWIKRGPTGVIGTNKKDAQDTVDT
Rv3106|M.tuberculosis_H37Rv SGTIPNVGGRINGSPN------EYVVGWIKRGPTGVIGTNKKDAQDTVDT
MLBr_00666|M.leprae_Br4923 SVTIPNTNGRVNGSRN------EYVVGWIKRGPTGVIGTNKKDSQDTVDT
MAV_4007|M.avium_104 TATIPNTGGRVEGSRN------EYVVGWIKRGPTGVIGTNKKDSQDTVDT
MAB_3482|M.abscessus_ATCC_1997 RGVILNEAGRIQGRDN------EYVVGWIKRGPVGVIGTNKSDSQETVDT
MSMEG_5681|M.smegmatis_MC2_155 ASVVPNDGGRVIDPATGAPVPGAYVAGWIKRGPTGFIGTNKSCAAQTVHH
: : *:: . . **.******* *.**:**. : :**
Mflv_4452|M.gilvum_PYR-GCK LLNDLSSAELA----DFGSDHPDKVQQWLQERQPKLITSEHWKLIDTHER
Mvan_1910|M.vanbaalenii_PYR-1 LLADLAAAELT----EFDAGHLEDIERWLLERQPKLVTNEHWKLIDAHER
TH_2151|M.thermoresistible__bu LLADLRDAELA----DFGPDHHDELAAWLSSRQPKLVTADHWNVIDEHER
MUL_2413|M.ulcerans_Agy99 LMADLTAAELN----TFPSSHTADLADWLSSRQPELVTAEHWKSIDHFER
MMAR_1526|M.marinum_M LMADLTAAELN----TFPSGHTADLADWLSSRQPELVTAEHWKSIDHFER
Mb3133|M.bovis_AF2122/97 LIKDLGNAKEGAECKSFPEDHADQVADWLAARQPKLVTSAHWQVIDAFER
Rv3106|M.tuberculosis_H37Rv LIKNLGNAKEGAECKSFPEDHADQVADWLAARQPKLVTSAHWQVIDAFER
MLBr_00666|M.leprae_Br4923 LMENLAGAND---TEKFGADHADRLAEWLAERQPKLVTSAHWQAIDRFER
MAV_4007|M.avium_104 LLADLANAGPDG-LADFPDDHADRLADWLASRQPKLVTTAHWELIDAHER
MAB_3482|M.abscessus_ATCC_1997 LLADLATAELR------DAPDVAALEEWLLGHEPHIVSQSDWLTIDSHER
MSMEG_5681|M.smegmatis_MC2_155 LVDDYNAGALTD-----PPRKPSALDKLVRARQPDVVDAAGWRAIDAAEI
*: : . . : : ::*.:: * ** *
Mflv_4452|M.gilvum_PYR-GCK TAGEGSGRPRVKVTSVEDLLRIG--HA--------------
Mvan_1910|M.vanbaalenii_PYR-1 SAGAGSGRPRVKLSNVAEMLRVG--GRG-------------
TH_2151|M.thermoresistible__bu GAGQQCGRPRVKLASVAELLRVG--HG--------------
MUL_2413|M.ulcerans_Agy99 AAGEALGRPRVKLPNVTEMLRIG--HA--------------
MMAR_1526|M.marinum_M AAGEALGRPRVKLPNVTEMLRIG--HA--------------
Mb3133|M.bovis_AF2122/97 AAGEPHGRPRVKLASLAELLRIG--LG--------------
Rv3106|M.tuberculosis_H37Rv AAGEPHGRPRVKLASLAELLRIG--LG--------------
MLBr_00666|M.leprae_Br4923 AAGEPHGRPRVKLPNLAELLRIG--HG--------------
MAV_4007|M.avium_104 AAGEARGRPRVKLPSLAKLLRVS--HG--------------
MAB_3482|M.abscessus_ATCC_1997 ATGEPQGRPRVKLETVPDLLAATGRHGEP------------
MSMEG_5681|M.smegmatis_MC2_155 ARG-GDDRPRDKFTTVPEMLAVAVTAPKPSVRQRLTAGLLG
* .*** *. .: .:*