For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPGAPSPSAPSIALNDENTMPVLGMGVAGLSDDETERAVSAALEIGCRLIDTAAAYGNEAAVGRALAASG IPRAELFVTTKVATADHGFTASREACKASLDRLGLDYVDLYLIHWPAPAVGKYVDAFGGLIQARGEGFTR SIGVSNFTEEHVSNVIDLTFVTPAVNQVELHPLLNQDELRKANAQHNVVTQSYTPLALGQLADNPTVTSI AGEYGKTPTQVLLRWNLQLGNAVIFGSSNAEHIVTNLDVFEFELASQHMDAINGLNDGTRLREDPMTFAG V*
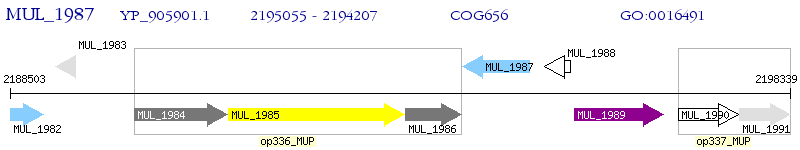
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1987 | - | - | 100% (282) | oxidoreductase |
| M. ulcerans Agy99 | MUL_2745 | - | 9e-12 | 29.00% (269) | hypothetical protein MUL_2745 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2996 | - | 1e-128 | 80.95% (273) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4205 | - | 1e-97 | 62.36% (271) | 2,5-didehydrogluconate reductase |
| M. tuberculosis H37Rv | Rv2971 | - | 1e-127 | 80.59% (273) | oxidoreductase |
| M. leprae Br4923 | MLBr_01669 | - | 1e-129 | 78.29% (281) | putative oxidoreductase of Aldo/keto reductase family |
| M. abscessus ATCC 19977 | MAB_1528c | - | 2e-64 | 46.10% (269) | oxidoreductase |
| M. marinum M | MMAR_1744 | - | 1e-159 | 98.93% (281) | oxidoreductase |
| M. avium 104 | MAV_3816 | - | 1e-121 | 79.31% (261) | 2,5-diketo-D-gluconic acid reductase A |
| M. smegmatis MC2 155 | MSMEG_2407 | - | 2e-98 | 62.36% (271) | morphine 6-dehydrogenase |
| M. thermoresistible (build 8) | TH_1364 | - | 1e-101 | 62.06% (282) | PROBABLE OXIDOREDUCTASE |
| M. vanbaalenii PYR-1 | Mvan_2161 | - | 3e-98 | 63.10% (271) | 2,5-didehydrogluconate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4205|M.gilvum_PYR-GCK MTS----AAAIPTVTLNDDNTMPVIGLGVGELSDAETEQAVTAALEAGYR
Mvan_2161|M.vanbaalenii_PYR-1 MTS----AAAIPTVILNDDNTMPVIGLGVGELSDAEAEQSVLAALEAGYR
MSMEG_2407|M.smegmatis_MC2_155 MTASHGQAAAIPTVTLNDDNTLPVVGIGVGELSDSEAERSVSAALEAGYR
TH_1364|M.thermoresistible__bu MTSSPGEAAATPTVTLNDDRSMPVLGLGVGELSDDEAERAVAAALEAGYR
MUL_1987|M.ulcerans_Agy99 MMPGAP-SPSAPSIALNDENTMPVLGMGVAGLSDDETERAVSAALEIGCR
MMAR_1744|M.marinum_M -MPGAP-SPSAPSIALNDENTMPVLGMGVAGLSDDETERAVSAALEIGCR
Mb2996|M.bovis_AF2122/97 MTGESG-AAAAPSITLNDEHTMPVLGLGVAELSDDETERAVSAALEIGCR
Rv2971|M.tuberculosis_H37Rv MTGESG-AAAAPSITLNDEHTMPVLGLGVAELSDDETERAVSAALEIGCR
MLBr_01669|M.leprae_Br4923 MMPSTP-RTSIPSITLNDENTMPLLGLGVAELSEDETERAVLAALEIGCR
MAV_3816|M.avium_104 ---------------------MPVLGLGVAELSDDETERAVSAALEVGCR
MAB_1528c|M.abscessus_ATCC_199 -------------MTLNSGTTIPQLGYGVWQVPDDQARAAVSAALQVGYR
:* :* ** :.: ::. :* ***: * *
Mflv_4205|M.gilvum_PYR-GCK LIDTAAAYGNEAAVGRAIAASGVPRDELFVTTKLATEDAGFQSSQDALRA
Mvan_2161|M.vanbaalenii_PYR-1 LIDTAAAYGNEAAVGRAIAKSGVPRGELFVTTKLATDDLGFQSSQDALRA
MSMEG_2407|M.smegmatis_MC2_155 LIDTAAAYGNEAAVGRAIAASGIPRDEIYVTTKLATPDQGFTSSQAAARA
TH_1364|M.thermoresistible__bu LIDTAASYGNEAGVGRAIAASGLPREEIFVTTKLATTDQGFQSSQDAVRA
MUL_1987|M.ulcerans_Agy99 LIDTAAAYGNEAAVGRALAASGIPRAELFVTTKVATADHGFTASREACKA
MMAR_1744|M.marinum_M LIDTAAAYGNEAAVGRAIAASGIPRAELFVTTKVATADHGFTASREACKA
Mb2996|M.bovis_AF2122/97 LIDTAYAYGNEAAVGRAIAASGVAREELFVTTKLATPDQGFTRSQEACRA
Rv2971|M.tuberculosis_H37Rv LIDTAYAYGNEAAVGRAIAASGVAREELFVTTKLATPDQGFTRSQEACRA
MLBr_01669|M.leprae_Br4923 LIDTAAAYGNEAAVSRAIAASGIPRAQLFVTTKLATPDQGFTKSQDACNA
MAV_3816|M.avium_104 LIDTAAAYGNEAAVGRAIAASGIPRAELFVTTKLATSDQGFKGAMDACEA
MAB_1528c|M.abscessus_ATCC_199 SIDTAKIYDNEAGTGAAIAESGIPRGDVFLTTKLWNSDQGYDNALRAFDA
**** *.***... *:* **:.* ::::***: . * *: : * *
Mflv_4205|M.gilvum_PYR-GCK SLERLGLDYVDLYLIHWPVSQQGKHVDSWGGLMKSKEVGLTRSIGVANFH
Mvan_2161|M.vanbaalenii_PYR-1 SLERLGLDYVDLYLIHWPAGSQGTYVDSWGGLMKLKELGLTRSIGVSNFH
MSMEG_2407|M.smegmatis_MC2_155 SLERLGLDYVDLYLIHWPGGDTSKYVDSWGGLMKVKEDGIARSIGVCNFG
TH_1364|M.thermoresistible__bu SLERLGLEYVDLYLIHWPAGEQGKYVDSWGGFLKIREDGLTRSIGVSNFD
MUL_1987|M.ulcerans_Agy99 SLDRLGLDYVDLYLIHWPAPAVGKYVDAFGGLIQARGEGFTRSIGVSNFT
MMAR_1744|M.marinum_M SLDRLGLDYVDLYLIHWPAPAAGKYVDAFGGLIQARGEGFTRSIGVSNFT
Mb2996|M.bovis_AF2122/97 SLDRLGLDYVDLYLIHWPAPPVGKYVDAWGGMIQSRGEGHARSIGVSNFT
Rv2971|M.tuberculosis_H37Rv SLDRLGLDYVDLYLIHWPAPPVGKYVDAWGGMIQSRGEGHARSIGVSNFT
MLBr_01669|M.leprae_Br4923 SLDRLGMDYVDLYLIHWPAPPVGQYVDAWGGMIQSRGEGHARSIGVCNFT
MAV_3816|M.avium_104 SLERLGLDYVDLYLIHWPAPALGTYVNSFGGMIQSRGNGHARSIGVSNFT
MAB_1528c|M.abscessus_ATCC_199 SLERLGTDYVDLYLIHWPVPELDEYVASFKALQRIQADGRAKAIGVSNFT
**:*** :********** . :* :: .: : : * :::***.**
Mflv_4205|M.gilvum_PYR-GCK GHHLDDIISLSFFTPAVNQIELHPLLNQAELRAVNAEHGIATEAYGPLGV
Mvan_2161|M.vanbaalenii_PYR-1 AQHLDDIIGLSFFTPAVNQIELHPLLNQAELRAVNAEHGIVTEAYSPLGV
MSMEG_2407|M.smegmatis_MC2_155 AEDLETIVSLTYFTPAVNQIELHPLLNQAALREVNAGYNIVTEAYGPLGV
TH_1364|M.thermoresistible__bu EEYLSDIIDLSFVTPAVNQIELHPLLNQADLRKVHAERNVLTQAYSPLGV
MUL_1987|M.ulcerans_Agy99 EEHVSNVIDLTFVTPAVNQVELHPLLNQDELRKANAQHNVVTQSYTPLAL
MMAR_1744|M.marinum_M EEHVSNVIDLTFVTPAVNQVELHPLLNQDELRKANAQHNVVTQSYTPLAL
Mb2996|M.bovis_AF2122/97 AEHIENLIDLTFVTPAVNQIELHPLLNQDELRKANAQHTVVTQSYCPLAL
Rv2971|M.tuberculosis_H37Rv AENIENLIDLTFVTPAVNQIELHPLLNQDELRKANAQHTVVTQSYCPLAL
MLBr_01669|M.leprae_Br4923 EEHLSAIIDLTFVTPAVNQIELHPLLNQDEMRKSNAQHNVITQSYTPLVL
MAV_3816|M.avium_104 EEYLTTVIDLTFVTPAVNQIELHPLLNQEALRKTNAEHNVVTQSYTPLAL
MAB_1528c|M.abscessus_ATCC_199 VDNLKRLIDETGEVPALNQIELHPRFTQPELRAFHAEYGIATEAWSPLGQ
. : ::. : .**:**:**** :.* :* :* : *::: **
Mflv_4205|M.gilvum_PYR-GCK GNLLSHSAVNAIAEAHGRTPAQVLIRWSLQLGNIVIPRSSSPERIRSNID
Mvan_2161|M.vanbaalenii_PYR-1 GSLLSNPAVTAIADAQDRTPAQVLIRWSLQLGNVVISRSSSPERIKSNLD
MSMEG_2407|M.smegmatis_MC2_155 GRLLDHPAVTAIAEAHGRTAAQVLLRWSIQLGNVVISRSANPERIASNLD
TH_1364|M.thermoresistible__bu GNLLADPTIGAVAQAHGKSPAQVLLRWNLQLGNAVVVRSSNPDRMRENLA
MUL_1987|M.ulcerans_Agy99 GQLADNPTVTSIAGEYGKTPTQVLLRWNLQLGNAVIFGSSNAEHIVTNLD
MMAR_1744|M.marinum_M GQLADNPTVTSIAGEYGKTPTQVLLRWNLQLGNAVIFGSSNAEHIATNLD
Mb2996|M.bovis_AF2122/97 GRLLDNPTVTSIASEYVKTPAQVLLRWNLQLGNAVVVRSARPERIASNFD
Rv2971|M.tuberculosis_H37Rv GRLLDNPTVTSIASEYVKTPAQVLLRWNLQLGNAVVVRSARPERIASNFD
MLBr_01669|M.leprae_Br4923 GRLMDNSTLTAIAAEYGKTPAQVLLRWNLQLGNAVVFRSAKAEHIASNFD
MAV_3816|M.avium_104 GKLNDHPTVNSVAAEYGKTASQVLLRWNLQLGNAVIFRSANAEHIASDFD
MAB_1528c|M.abscessus_ATCC_199 GTILEDATIGAIAQAHDVSAAQVILRWHLQLGNVVIPKSVTPARIAANFD
* : ..:: ::* :.:**::** :**** *: * . :: ::
Mflv_4205|M.gilvum_PYR-GCK VFGFELSEEQMSTLNGLDDG-TRFRPDPETYSGS----------------
Mvan_2161|M.vanbaalenii_PYR-1 VFDFELTADQMAALDGLDEG-TRFRPDPETYTGS----------------
MSMEG_2407|M.smegmatis_MC2_155 VFGFELTADEMETLNGLDDG-TRFRPDPATYTGS----------------
TH_1364|M.thermoresistible__bu VFDFELTDDEMNTINGMDNG-TRYRPDPXXGEGAAAEHQSADSAAGTAVP
MUL_1987|M.ulcerans_Agy99 VFEFELASQHMDAINGLNDG-TRLREDPMTFAGV----------------
MMAR_1744|M.marinum_M VFEFELASQHMDAINGLNDG-TRLREDPMTFAGV----------------
Mb2996|M.bovis_AF2122/97 VFDFELAAEHMDALGGLNDG-TRVREDPLTYAGT----------------
Rv2971|M.tuberculosis_H37Rv VFDFELAAEHMDALGGLNDG-TRVREDPLTYAGT----------------
MLBr_01669|M.leprae_Br4923 VFDFELAVNHMDAMNELHDG-TRLRPDPETYAGS----------------
MAV_3816|M.avium_104 VFDFELAAEHMDAINALNDG-TRLRPDPDTYEGS----------------
MAB_1528c|M.abscessus_ATCC_199 VFGFELSTDEVERITALDASDGRIGPDPTTFNLH----------------
** ***: :.: : :. . * **
Mflv_4205|M.gilvum_PYR-GCK ----------------------------------------
Mvan_2161|M.vanbaalenii_PYR-1 ----------------------------------------
MSMEG_2407|M.smegmatis_MC2_155 ----------------------------------------
TH_1364|M.thermoresistible__bu GHPVRRGGCARRQRRPGTAADHLLHRRSDGLSVEQVDHQR
MUL_1987|M.ulcerans_Agy99 ----------------------------------------
MMAR_1744|M.marinum_M ----------------------------------------
Mb2996|M.bovis_AF2122/97 ----------------------------------------
Rv2971|M.tuberculosis_H37Rv ----------------------------------------
MLBr_01669|M.leprae_Br4923 ----------------------------------------
MAV_3816|M.avium_104 ----------------------------------------
MAB_1528c|M.abscessus_ATCC_199 ----------------------------------------