For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTRSGRRALLVGLTAASVGVLYGYDLS AIAGALLSLSEEFELTTREQELLTTTAVLGQIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVS VPMLVVARLLLGVTIGLSVVVVPVYVAESAPAAVRGSLVTAYQLATLSGIVVGYLVGYLLAGSHGWRAMF GLAAAPATLLLPLLWRMPDTARWYLLKGRIADARSALRRIQPEADIDAELADMAAAVDERGGGIGEMVRR PYLRATLFVIALGFLVQITGINAIIYYSPRLFAAMGFAGYFAMLALPAMVQVAGLAAVCASLFLVDRLGR RPILLSGIATMITADAVLITVFANDSDGGTGLVLGFAGVLLFIIGFNFGFGSLVWVYAAESFPSRLRSMG SSPMLTSTLTANAIVAAFSLTMLRVLGGAGVFAVFGTFAVVAFVVVYRFAPETKGRKLEEIRHFWENGGR WPAERSPAADEP
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
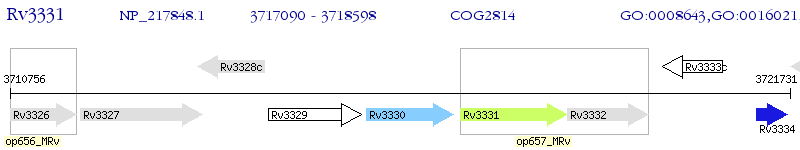
| M. tuberculosis H37Rv | Rv3331 | sugI | - | 100% (502) | PROBABLE SUGAR-TRANSPORT INTEGRAL MEMBRANE PROTEIN SUGI |
| M. tuberculosis H37Rv | Rv1672c | - | 6e-13 | 26.53% (294) | integral membrane transport protein |
| M. tuberculosis H37Rv | Rv1902c | nanT | 4e-12 | 26.15% (195) | sialic acid-transport integral membrane protein NanT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3364 | sugI | 0.0 | 99.80% (502) | sugar-transport integral membrane protein SugI |
| M. gilvum PYR-GCK | Mflv_3055 | - | 1e-50 | 27.82% (478) | sugar transporter |
| M. leprae Br4923 | MLBr_01562 | - | 1e-07 | 27.35% (223) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_0365c | - | 1e-142 | 55.48% (456) | sugar-transport integral membrane protein |
| M. marinum M | MMAR_1191 | sugI | 1e-177 | 68.44% (450) | sugar-transport integral membrane protein SugI |
| M. avium 104 | MAV_4306 | - | 1e-176 | 69.69% (452) | transporter, major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_5559 | - | 1e-56 | 31.84% (468) | metabolite/sugar transport protein |
| M. thermoresistible (build 8) | TH_2640 | - | 5e-27 | 27.10% (321) | PUTATIVE sugar transporter |
| M. ulcerans Agy99 | MUL_1444 | sugI | 1e-178 | 68.89% (450) | sugar-transport integral membrane protein SugI |
| M. vanbaalenii PYR-1 | Mvan_3472 | - | 3e-50 | 28.90% (474) | sugar transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3331|M.tuberculosis_H37Rv ----MTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTR
Mb3364|M.bovis_AF2122/97 ----MTTLWQPHRNDYSPIPGRGVHARRGARRPRPRGGRAERPGTGQLTR
MMAR_1191|M.marinum_M --------------------------------------------------
MUL_1444|M.ulcerans_Agy99 --------------------------------------------------
MAV_4306|M.avium_104 -----------------------------------------------MAR
MAB_0365c|M.abscessus_ATCC_199 -----------------------------------------MSLITDLRS
Mflv_3055|M.gilvum_PYR-GCK ----------------------------MAGQGGPAGESPELYEKGQDFS
Mvan_3472|M.vanbaalenii_PYR-1 ----------------------------MAGQGGPVGDGPEIFEQGQEFS
TH_2640|M.thermoresistible__bu --------------------------------------------------
MSMEG_5559|M.smegmatis_MC2_155 -------------------------------MSTDTQDSADARAKTPGQL
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Rv3331|M.tuberculosis_H37Rv SGRRALLVGLTAASVGVLYGYDLSAIAGALLSLSEEFELTTREQELLTTT
Mb3364|M.bovis_AF2122/97 SGRRALLVGLTAASVGVLYGYDLSAIAGALLSLSEEFELTTREQELLTTT
MMAR_1191|M.marinum_M -MQRGILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTM
MUL_1444|M.ulcerans_Agy99 -MQRGILVALTAASVGLIYGYDLSIIAGAQLFITEEFGLSTHQQELLTTM
MAV_4306|M.avium_104 GSRRGLLVGLTAASVGVIYGYDLSIIAGAQLFVTEDFGLSTRQQELLTTM
MAB_0365c|M.abscessus_ATCC_199 TSRLGVMVAVTAAAVGVIYGYDSSNIAGALLFLTEDLHLSTHDQQVAATA
Mflv_3055|M.gilvum_PYR-GCK SGKTAIRIASVAALGGLLFGYDSAVINGAVASIQEDFGIGNYALGLAVAS
Mvan_3472|M.vanbaalenii_PYR-1 SGKTAVRIASVAALGGLLFGYDSAVINGAVDSIQEDFGIGNAELGFAVAS
TH_2640|M.thermoresistible__bu --------------------------------------------------
MSMEG_5559|M.smegmatis_MC2_155 TG-AVVIVALVSAVSGMLYGYDTGIISGALLQITKDFSIAEAWKQVIAAS
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
Rv3331|M.tuberculosis_H37Rv AVLGQIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVSVPMLVV
Mb3364|M.bovis_AF2122/97 AVLGQIAGALGGGILANAIGRKKSVVLIVAGYAVFALLGATSVSVPMLVV
MMAR_1191|M.marinum_M VVIGQIVGALGAGVLANAIGRKKSVVMLLVAYTMFAVLGSLSVSLPMLLA
MUL_1444|M.ulcerans_Agy99 VVIGQIVGALGAGVLANAIGRKKSVVMLLVAYTMFAVLGALSVSLPMLLA
MAV_4306|M.avium_104 AVIGQIGGALFAGVLANAIGRRRSVLLILSGYAVFALLAAFSVGLPMLLT
MAB_0365c|M.abscessus_ATCC_199 VVIGEIFGALIGGPLSNKIGRKRSMVLVAATFGIFSLLSALAVDLNTLAA
Mflv_3055|M.gilvum_PYR-GCK ALLGAAVGALSAGRIADRIGRIAVMKIAAVLFLLSAFGTAFAPETITLVI
Mvan_3472|M.vanbaalenii_PYR-1 ALLGAAAGAMTAGRIADRIGRIAVMKIAAVLFLVSAFGTGFAHEVWAVVL
TH_2640|M.thermoresistible__bu --------------------------------------------------
MSMEG_5559|M.smegmatis_MC2_155 ILLGAVIGALVCSHLSQKRGRKGTLLMLAVVFVIGSLWCAISPNPVLLSV
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
Rv3331|M.tuberculosis_H37Rv ARLLLGVTIGLSVVVVPVYVAESAPAA-VRGSLVTAYQLATLSGIVVGYL
Mb3364|M.bovis_AF2122/97 ARLLLGVTIGLSVVVVPVYVAESAPAA-VRGSLVTAYQLATLSGIVVGYL
MMAR_1191|M.marinum_M ARFLLGLAVGVSIVVVPVYVAESAPAA-VRGSLLTVYQLTTVSGLIVGYL
MUL_1444|M.ulcerans_Agy99 ARFLLGLAVGVSIVVVPVYVAESAPAA-VRGSLLTVYQLTTVSGLIVGYL
MAV_4306|M.avium_104 ARLLLGLTIGVTVVVVPVYVAESAPTA-VRGALLTAYQLAIVSGLIVGYL
MAB_0365c|M.abscessus_ATCC_199 ARFLLGLTVGVSVVVVPVFVAESSPTK-IRGSMLVLYQLACVTGIIAGYL
Mflv_3055|M.gilvum_PYR-GCK GRIVGGVGVGVASVIAPAYIAETSPPG-IRGRLGSLQQLAIVSGIFASFA
Mvan_3472|M.vanbaalenii_PYR-1 FRIVGGIGVGVASVIAPAYIAETSPPG-IRGRLGSLQQLAIVSGIFASFA
TH_2640|M.thermoresistible__bu --------------------------------------LAIVSGIFLSLL
MSMEG_5559|M.smegmatis_MC2_155 GRLVLGFAVGGATQTAPMYVAELSPPK-YRGRLVLCFQIAIGVGIVIATI
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
* . .
Rv3331|M.tuberculosis_H37Rv VGYLLAG------------SHGWRAMFGLAAAPATLLL--------PLLW
Mb3364|M.bovis_AF2122/97 VGYLLAG------------SHGWRAMFGLAAAPATLLL--------PLLW
MMAR_1191|M.marinum_M TGYLLAG------------THSWRWMLGLATVPAMLLL--------PLLI
MUL_1444|M.ulcerans_Agy99 TGYLLAG------------THSWRWMLGLATVPAMLLL--------PLLI
MAV_4306|M.avium_104 SGYLLAG------------THSWRWMLGLACVPAVLLL--------PLVF
MAB_0365c|M.abscessus_ATCC_199 IAWALSS------------TGSWRLMLGLAAVPAALVL--------IALL
Mflv_3055|M.gilvum_PYR-GCK VNYLLQWLAGGPNEPLWLGMDAWRWMFLAMAVPAVVYG--------GLAF
Mvan_3472|M.vanbaalenii_PYR-1 VNWLLQWAAGGPNEPLWFGLDAWRWMFLAMALPAVLYG--------VLAF
TH_2640|M.thermoresistible__bu IDGILAALAGGSREELWLNMEAWRWMFLMMAVPAVLYG--------ALTF
MSMEG_5559|M.smegmatis_MC2_155 VGAS--------------EHIPWRWSIGAAAVPAAIML--------VLLL
MLBr_01562|M.leprae_Br4923 VGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAGAILA
:
Rv3331|M.tuberculosis_H37Rv RMPDTARWYLLKGRIADARSALRRIQPE-ADIDAELADMAAAV-DERGG-
Mb3364|M.bovis_AF2122/97 RMPDTARWYLLKGRIADARSALRRIQPE-ADIDAELADMAAAV-DERGG-
MMAR_1191|M.marinum_M RMPDTPRWYVMKGRIQEARAALLRVDPA-ADVEEELAEIGTAL-SEGSG-
MUL_1444|M.ulcerans_Agy99 RMPDTPRWYVMKGRIQEARAALLRVDPA-ADVEEELAEIGTAL-SEGSG-
MAV_4306|M.avium_104 RMPDTARWYLLKGRVDDARRALLRVEPA-ARVDDELAEIGRAV-SEEAAS
MAB_0365c|M.abscessus_ATCC_199 KLPDTARWYMMRGDRARAREVLSMVDRD-VDIDHELDEIAEALRAEGGGS
Mflv_3055|M.gilvum_PYR-GCK TIPESPRYLVATHKIPEARRVLSMLLGQ-KNLEITITRIRETLEREDRPS
Mvan_3472|M.vanbaalenii_PYR-1 TIPESPRYLVASHKIPEARRVLSMLLGK-KNLEITITRIRETLEREDKPS
TH_2640|M.thermoresistible__bu TIPESPRYLVATHRVPEARRVLSRLLGA-KNLEITINRIERSLRAEKPPS
MSMEG_5559|M.smegmatis_MC2_155 RLPESPRWLIKDGDPDKAREVLERVRPDGYDIDGELDEMTTLVRKEQTAK
MLBr_01562|M.leprae_Br4923 TLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENPVV
:. :. . * : : : *
Rv3331|M.tuberculosis_H37Rv ---GIGEMVRRPYLRATLFVIALGFLVQITG---INAIIYYSPRLFAAMG
Mb3364|M.bovis_AF2122/97 ---GIGEMVRRPYLRATLFVIALGFLVQITG---INAIIYYSPRLFAAMG
MMAR_1191|M.marinum_M ---GVSEMLRPPFLRATIFVITLGFLIQITG---INAIIYYSPRIFEAMG
MUL_1444|M.ulcerans_Agy99 ---GVSEMLRPPFLRATIFVITLGFLIQITG---INAIIYYSPRIFEAMG
MAV_4306|M.avium_104 LPAMLAEMVRSPYRRATVFVVVLGFLIQITG---INAIIYYSPRIFEAMG
MAB_0365c|M.abscessus_ATCC_199 VRARLREMVTAPYRRATFFVVGLGFFIQITG---INAVVYYGPRIFEAMG
Mflv_3055|M.gilvum_PYR-GCK -WRDLKKPTGGIYG-IVWVGLGLSIFQQFVG---INVIFYYSNVLWQAVG
Mvan_3472|M.vanbaalenii_PYR-1 -WRDMRKPTGGLYG-IVWVGLGLSIFQQFVG---INVIFYYSNVLWQAVG
TH_2640|M.thermoresistible__bu -WSDLRKPTGGMYG-IVWVGLGLSIFQQFVG---INVIFYYSNVLWEAVG
MSMEG_5559|M.smegmatis_MC2_155 -TRGWPGLRAAWVRPALILGCGIAIFTQLSG---IEMIIYYSPTILTDNG
MLBr_01562|M.leprae_Br4923 PFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYS-ALRAGIG
. : : . *: :. *. : *
Rv3331|M.tuberculosis_H37Rv FAGYFAMLALPAMVQVAGLAAVCASLFLVDR-LGRRPILLSGIATMITAD
Mb3364|M.bovis_AF2122/97 FAGYFAMLALPAMVQVAGLAAVCASLFLVDR-LGRRPILLSGIATMITAD
MMAR_1191|M.marinum_M FTGDFALLGLPALVQIAGVAAVITSLLLVDR-VGRRPILLSGIAIMIVAD
MUL_1444|M.ulcerans_Agy99 FTGDFALLGLPALVQIAGVAAVITSLLLVDR-VGRRPILLSGIAIMIVAD
MAV_4306|M.avium_104 FTGNFALLALPALVQVAGLVAVGTALLLVDR-VGRRPILLCGTAMMIVAD
MAB_0365c|M.abscessus_ATCC_199 MSGYFAKLGLPALVQVAALLAVFISMSSIDR-MGRRPILMIGIGIMIVAD
Mflv_3055|M.gilvum_PYR-GCK FS-ADESAVYTVITSVVNVLTTLIAIALIDK-IGRKPLLLIGSVGMAVTL
Mvan_3472|M.vanbaalenii_PYR-1 FS-ADESAVYTVITSVINVLTTLIAIALIDK-IGRKPLLLIGSSGMAVTL
TH_2640|M.thermoresistible__bu FD-ESQSFLITVITSVTNIVTTLIAIALIDK-IGRKPLLLIGSTGMAGTL
MSMEG_5559|M.smegmatis_MC2_155 FS-ESVALQVSVALGVSYLVAQLVGLSIIDK-VGRRRLTLIMIPGAAVSL
MLBr_01562|M.leprae_Br4923 FIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRGV
: : : :. :
Rv3331|M.tuberculosis_H37Rv AVLITVFANDSD------------GGTGLVLGFAGVLLFIIGFNFGFGSL
Mb3364|M.bovis_AF2122/97 AVLITVFANDSD------------GGTGLVLGFAGVLLFIIGFNFGFGSL
MMAR_1191|M.marinum_M VALMAVFAR---------------GQGAAILGFAGILLFIIGYTMGFGSL
MUL_1444|M.ulcerans_Agy99 VALMAVFAR---------------GQGAAILGFAGILLFIIGYTMGFGSL
MAV_4306|M.avium_104 VVLVAVFGR---------------GPGGVIAGFAGVLLFIFGYTMGFGSL
MAB_0365c|M.abscessus_ATCC_199 ALLVGVFAVGGSS----------FGGLLTFLGFLGIVLIAVGFTFGFGSL
Mflv_3055|M.gilvum_PYR-GCK STMAFIFANAELIENEAGEMVPSLPGASGVIALIAANLFVVAFGMSWGPV
Mvan_3472|M.vanbaalenii_PYR-1 ITMAVIFANATVGADGN----PSLPGASGVIALIAANLFVVAFGMSWGPV
TH_2640|M.thermoresistible__bu GVMAVIFGTAPVIDGQP-----QLGDVAGPVALVAANLFVVSFGMSWGPV
MSMEG_5559|M.smegmatis_MC2_155 FVLGTLFATG------------HSGRDSVPFIVACLVVFMLFNAGGLQLM
MLBr_01562|M.leprae_Br4923 PYFPNLLAPIVVGG-------IGIGMAVVPLTLSAIAGVGFDRIGPVSAI
: ::. . . . :
Rv3331|M.tuberculosis_H37Rv VWVYAAESFPSRLRSMGSSPMLTSTLTANAIVAAFSLTMLRVLGGAGVFA
Mb3364|M.bovis_AF2122/97 VWVYAAESFPSRLRSMGSSLMLTSTLTANAIVAAFSLTMLRVLGGAGVFA
MMAR_1191|M.marinum_M GWVYASESFPTRLRSIGSSTMLTANLIANAIVAGVFLTMLHSLGGSGAFA
MUL_1444|M.ulcerans_Agy99 GWVYASESFPTRLRSIGSSTMLTANLIANAIVAAVFLTMLHSLGGSGAFA
MAV_4306|M.avium_104 GWVYASESFPSRLRSIGSSTMLTSNLVANAIVAAVFLTLLHSLGGAGTFA
MAB_0365c|M.abscessus_ATCC_199 VWVYAGESFPARLRSYGASAMLTSDLVANAVVSMYFLTLLTALGGVVAFG
Mflv_3055|M.gilvum_PYR-GCK VWVLLGEMFPNRIRAAALGLAAAGQWTANWVITVSFPELRN--FLGAAYG
Mvan_3472|M.vanbaalenii_PYR-1 VWVLLGEMFPNRIRAAALGLAAAGQWAANWLITVTFPGLRE--HLGLAYG
TH_2640|M.thermoresistible__bu VWVLLGEMFPNRIRAAALGLAAAGQWTANWVITVTFPGLRE--HLGPAYG
MSMEG_5559|M.smegmatis_MC2_155 GWLTGSETYPLAVRPAGTAAQSAALWGTNLLITLTLLTLISGIGVGPAMW
MLBr_01562|M.leprae_Br4923 ALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGY-
: . * : . .
Rv3331|M.tuberculosis_H37Rv VFGTFAVVAFVVVYRFAPETKGRKLEEIRHFWENGGRWPAERSPAADEP
Mb3364|M.bovis_AF2122/97 VFGTFAVVAFVVVYRFAPETKGRKLEEIRHFWENGGRWPAERSPAADEP
MMAR_1191|M.marinum_M VFGVLALVAFGVVYRYAPETKGRQLEEIRHFWENGGRWPEKLTEAP---
MUL_1444|M.ulcerans_Agy99 VFGVLALVAFGVVYRYAPETKGRQLEEIRHFWENGGRWPEKLTEAP---
MAV_4306|M.avium_104 VFAVLAVVAFAFVHRYAPETKGRQLEDIRHFWENGGRWD----------
MAB_0365c|M.abscessus_ATCC_199 VFGVLATAALVFVHVLAPETKGRNLEEIRHFWESGGTWPET--------
Mflv_3055|M.gilvum_PYR-GCK FYALCAVLSGLFVWKWVRETKGVSLEDMHGEILHGDKS-ATG-------
Mvan_3472|M.vanbaalenii_PYR-1 FYGLCAVLSGLFVWRWVMETKGVSLEDMHGEILHADKT-ATG-------
TH_2640|M.thermoresistible__bu FYALCAVLSLLFVWRWVAETKGRTLEDMQPDVPGHDRASAPG-------
MSMEG_5559|M.smegmatis_MC2_155 LYGLFNVAAWLFVYFRMPDLTGRSLEEIEGKLRDGKFRPADFAR-----
MLBr_01562|M.leprae_Br4923 TYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL------
:. . .: * *::