For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSLERDGHVLLIGLNRPQKRNSFDRAMFADLSRAYALLETDDSVRAGVLFAHGDHFTGGLDLVDVAPSIA AGQSPFPADGRDPWRLDGPWRTPLIAVAHGWCMTLGIELLLASDIRIAAAGTRFSQLEVQRGIYPFGGAT IRLTRDAGWGNAMRWLLTGDEFDAAEAHRIGLVQEVADDAATALTRAREIAQTIADRAAPLGVRATLASV HLARREGEAAAIERLVPDIRALFTSADAAEGVQSFIERRQARFTGR
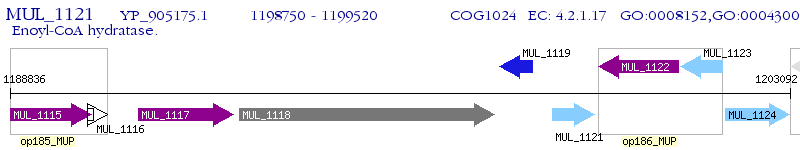
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1121 | - | - | 100% (256) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_4076 | echA19 | 1e-32 | 35.63% (261) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 3e-29 | 34.48% (261) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3545 | echA19 | 2e-30 | 34.34% (265) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_1542 | - | 5e-30 | 33.60% (253) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv3516 | echA19 | 1e-29 | 33.96% (265) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 3e-27 | 34.50% (258) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1493c | - | 5e-73 | 57.25% (255) | enoyl-CoA hydratase |
| M. marinum M | MMAR_0471 | - | 1e-144 | 99.61% (256) | enoyl-CoA hydratase |
| M. avium 104 | MAV_0643 | - | 2e-31 | 34.48% (261) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_3362 | - | 4e-74 | 56.03% (257) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_3131 | - | 8e-30 | 32.81% (256) | PUTATIVE putative enoyl-CoA hydratase EchA1 |
| M. vanbaalenii PYR-1 | Mvan_5216 | - | 1e-29 | 33.59% (256) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1542|M.gilvum_PYR-GCK MLHFQTSLWHRRWKADQFVSDEQNPEKAPDALVEQRGHTLIVTMNRPEKR
Mvan_5216|M.vanbaalenii_PYR-1 ------------------MSD---PQKGPDALVEQRGHTLIVTLNRPEAR
Mb3545|M.bovis_AF2122/97 ------------------------MESGPDALVERRGHTLIVTMNRPAAR
Rv3516|M.tuberculosis_H37Rv ------------------------MESGPDALVERRGHTLIVTMNRPAAR
MAV_0643|M.avium_104 -----------------MAEASANEKTGPDALVEQRGHTLIVTLNRPHAR
MLBr_02402|M.leprae_Br4923 -------------------------MKYDTILVDGYQRVGIITLNRPQAL
TH_3131|M.thermoresistible__bu ----------------------------MIVEVRRQDRILVIAMNRPAKR
MUL_1121|M.ulcerans_Agy99 ------------------------------MSLERDGHVLLIGLNRPQKR
MMAR_0471|M.marinum_M ---------------MQMTTTETSLPKLATVSLERDGHVLLIGLNRPQKR
MAB_1493c|M.abscessus_ATCC_199 ------------------------MSDYETLRIRRDGYVLVIGLNRPAKR
MSMEG_3362|M.smegmatis_MC2_155 ----------------------MSFDRYETLAVTRDGAVLVIGLNRPDKR
: :: :***
Mflv_1542|M.gilvum_PYR-GCK NALSAEMMQIMVEAWDRVDSDPEIRSCILTGAGGAFCAGMDLKKADSQAP
Mvan_5216|M.vanbaalenii_PYR-1 NALSTEMLSIMVDAWNRVDEDPEIRTCILTGAGGYFCAGMDLKAASKKPP
Mb3545|M.bovis_AF2122/97 NALSTEMMRIMVQAWDRVDNDPDIRCCILTGAGGYFCAGMDLKAATQKPP
Rv3516|M.tuberculosis_H37Rv NALSTEMMRIMVQAWDRVDNDPDIRCCILTGAGGYFCAGMDLKAATQKPP
MAV_0643|M.avium_104 NALSTEMMEIMVQAWDRVDNDDDIRCCILTGAGGYFCAGMDLKAATKKPP
MLBr_02402|M.leprae_Br4923 NALNSQMMNEITNAAKELDIDPDVGAILITGSPKVFAAGADIKEMASLTF
TH_3131|M.thermoresistible__bu NAVNKEMAEALSAALDLMDDDDELWAGVLTGTREFFCAGTDLKDGADART
MUL_1121|M.ulcerans_Agy99 NSFDRAMFADLSRAYALLETDDSVRAGVLFAHGDHFTGGLDLVDVAPSIA
MMAR_0471|M.marinum_M NSFDRAMFADLSRAYALLETDDSVRAGVLFAHGDHFTGGLDLVDVAPSIA
MAB_1493c|M.abscessus_ATCC_199 NAFDKTMLEELALALGEYETDTDLRAAVLYGEGPLFTAGLDLASVAAEIQ
MSMEG_3362|M.smegmatis_MC2_155 NAFNREMIGELAEVLGKLDSDDELRVGVLYGEGKGFTSGLDLPDVAPSLQ
*:.. * : . : * .: :: . * .* *:
Mflv_1542|M.gilvum_PYR-GCK GDSFKDGSYDPSKIPALLKGRRLKKPLIAAVEGAAIAGGTEILQGTDIRV
Mvan_5216|M.vanbaalenii_PYR-1 GDSFADGSYDPSKIEGLLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRV
Mb3545|M.bovis_AF2122/97 GDSFKDGSYDPSRIDALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
Rv3516|M.tuberculosis_H37Rv GDSFKDGSYGPSRIDALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
MAV_0643|M.avium_104 GESFKDGSYDPSRIDALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
MLBr_02402|M.leprae_Br4923 TDAFDADFFS-----AWGKLAAVRTPMIAAVAGYALGGGCELAMMCDLLI
TH_3131|M.thermoresistible__bu ERGGEYGIIR----------RRRRKPLIAAVEGACFGGGMEIAFACDLVV
MUL_1121|M.ulcerans_Agy99 AGQSP--FPADGRDPWRLDG-PWRTPLIAVAHGWCMTLGIELLLASDIRI
MMAR_0471|M.marinum_M AGQSP--FPADGRDPWRLDG-PWRTPLIAVAHGWCMTLGIELLLASDIRI
MAB_1493c|M.abscessus_ATCC_199 GGASL--TPEGGINPWQVDGRQLSKPLLVAVHGKVLTLGIELALAADIVI
MSMEG_3362|M.smegmatis_MC2_155 DGSLQ--IPEGGINPWQVDGRRLTKPLVTAVHGKCLTLGIELLLASDIVV
.*::... * : * *: *: :
Mflv_1542|M.gilvum_PYR-GCK AGESAKFGVSEAKWSLYPMGGSAVRLVRQIPYTVACDILLTGRHISAAEA
Mvan_5216|M.vanbaalenii_PYR-1 AGESAKFGISEAKWSLYPMGGSAVRLPRQIPYTIACDLLLTGRHITAAEA
Mb3545|M.bovis_AF2122/97 AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTLACDLLLTGRHITAAEA
Rv3516|M.tuberculosis_H37Rv AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTLACDLLLTGRHITAAEA
MAV_0643|M.avium_104 AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEA
MLBr_02402|M.leprae_Br4923 AADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEA
TH_3131|M.thermoresistible__bu AATTARFALPESRRGLVPSSGALLRATRSLPIHLAKELMITGAELSGERA
MUL_1121|M.ulcerans_Agy99 AAAGTRFSQLEVQRGIYPFGGATIRLTRDAGWGNAMRWLLTGDEFDAAEA
MMAR_0471|M.marinum_M AAAGTRFSQLEVQRGIYPFGGATIRLTRDAGWGNAMRWLLTGDEFDAAEA
MAB_1493c|M.abscessus_ATCC_199 ADETATFAQLEVNRGIYPFGGATIRFPRTAGWGNAMRWMLTADTFDAVEA
MSMEG_3362|M.smegmatis_MC2_155 ASESATFAQLEIARGIYPFGGATPRFPRVAGWGNAMRWMLTAEEFDSAEA
* : *. * .: * *. * * * ::*. : . .*
Mflv_1542|M.gilvum_PYR-GCK KEFGLIGHVVPDGQALTK-ALEIAEVING-NGPLAVQAILKTIRETEGMH
Mvan_5216|M.vanbaalenii_PYR-1 LQYGLIGHVVPDGTALDK-ALEIAEVINN-NGPLAVQAILKTIRETEGMH
Mb3545|M.bovis_AF2122/97 KEMGLIGHVVPDGQALTK-ALELADAISA-NGPLAVQAILRSIRETECMP
Rv3516|M.tuberculosis_H37Rv KEMGLIGHVVPDGQALTK-ALELADAISA-NGPLAVQAILRSIRETECMP
MAV_0643|M.avium_104 KEMGLIGHVVPDGQALAK-ALEIAEVIEN-NGPLAVQAILRAIRETEGMH
MLBr_02402|M.leprae_Br4923 ERSGLVSRVVLADDLLPE-AKAVATTISQ-MSRSATRMAKEAVNRSFEST
TH_3131|M.thermoresistible__bu HHFGLVNRIAEPGAALEA-AIDLAEDVCQ-SSPTAVQAILEALADQLDEA
MUL_1121|M.ulcerans_Agy99 HRIGLVQEVADDAATALTRAREIAQTIADRAAPLGVRATLASVHLARREG
MMAR_0471|M.marinum_M HRIGLVQEVADDAATALTRAREIAQTIADRAAPLGVRATLASAHLARREG
MAB_1493c|M.abscessus_ATCC_199 HRIGIVQEIVPVGEHVDT-AIAIAQTIAR-QAPLGVQATLRNARLAVREG
MSMEG_3362|M.smegmatis_MC2_155 LRIGVVQEVVDDGAHIAR-AVQLAHVIAA-QAPLGVQATLRSARLAQRDG
. *:: .:. * :* : . ..:
Mflv_1542|M.gilvum_PYR-GCK EEEAFKPDTANGIPVFLSEDSKEGPKAFLEKRKPVWKLK
Mvan_5216|M.vanbaalenii_PYR-1 EEEAFKPDTANGIPVFLSEDAKEGPRAFKEKRAPNFQMK
Mb3545|M.bovis_AF2122/97 ENEAFKIDTQIGIKVFLSDDAKEGPRAFAEKRAPNFQNR
Rv3516|M.tuberculosis_H37Rv ENEAFKIDTQIGIKVFLSDDAKEGPRAFAEKRAPNFQNR
MAV_0643|M.avium_104 ENEAFKIDTQIGIKVFLSEDAKEGPRAFAEKRKPNFQNR
MLBr_02402|M.leprae_Br4923 LAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR
TH_3131|M.thermoresistible__bu DAVGWTATAKAVDRVVSSDDLQEGLAAFFERRPPRWVGR
MUL_1121|M.ulcerans_Agy99 EAAAIERLVPDIRALFTSADAAEGVQSFIERRQARFTGR
MMAR_0471|M.marinum_M EAAAIERLVPDIRALFTSADAAEGVQSFIERRQARFTGR
MAB_1493c|M.abscessus_ATCC_199 DAAAEEQLVPTVRELFTSEDATLGVQAFLSRTTAEFVGR
MSMEG_3362|M.smegmatis_MC2_155 EAAAEAELVPTIVKLFGTEDSRIGLESFATRRPAQFVGR
. . : * * :* : . : :