For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AHESRAGRRPSTTKRHIADVAIDLFAARTFAEVSVDDVAQAAGIARRTLFRYYASKNAIPWGDFDTHLAQ LQDLLERIDGHVRLGKALREALLAFNTYDESETIRHRQRMRIILQTAELQAYSMTMYAGWRAVIAGFVAR RLSVKPTDLVPQTVAWTMLGVALSAYEHWLSDESVSLPEALGNAFDVVGAGLEKLD*
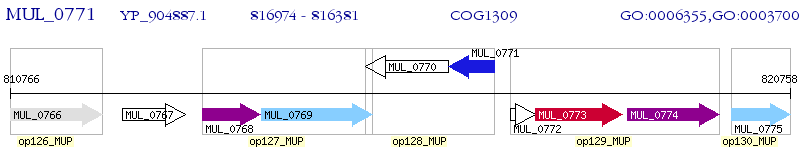
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0771 | - | - | 100% (197) | transcriptional regulator |
| M. ulcerans Agy99 | MUL_2933 | - | 1e-10 | 26.32% (190) | TetR family transcriptional regulator |
| M. ulcerans Agy99 | MUL_4892 | - | 3e-08 | 26.97% (178) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0710c | - | 2e-85 | 78.68% (197) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_5062 | - | 5e-67 | 64.80% (196) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0691c | - | 2e-85 | 78.68% (197) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00064 | - | 3e-06 | 43.10% (58) | putative transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3837 | - | 3e-60 | 59.49% (195) | transcriptional regulatory protein TetR |
| M. marinum M | MMAR_1019 | - | 1e-109 | 100.00% (197) | transcriptional regulator |
| M. avium 104 | MAV_4481 | - | 5e-86 | 80.41% (194) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1420 | - | 5e-73 | 70.41% (196) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_1899 | - | 1e-67 | 70.62% (177) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_1294 | - | 2e-70 | 65.82% (196) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5062|M.gilvum_PYR-GCK ----MSAARPRVGRRRSTSWE------HISDVAIDLFMARGFDDVSVDDV
Mvan_1294|M.vanbaalenii_PYR-1 ----MSAAKARVGRRRSTSWE------HLSDVAIDLFMARGFDDVSVDDV
TH_1899|M.thermoresistible__bu -------------------------------VALDLFADRGFDEVSVDDV
MUL_0771|M.ulcerans_Agy99 -----MAHESRAGRRPSTTKR------HIADVAIDLFAARTFAEVSVDDV
MMAR_1019|M.marinum_M -----MAHESRAGRRPSTTKR------HIADVAIDLFAARTFAEVSVDDV
Mb0710c|M.bovis_AF2122/97 -----MPHESRVGRRRSTTPH------HISDVAIELFAAHGFTDVSVDDI
Rv0691c|M.tuberculosis_H37Rv -----MPHESRVGRRRSTTPH------HISDVAIELFAAHGFTDVSVDDI
MAV_4481|M.avium_104 -----MPQQ-RVGRRRSTTPH------HITDVAIDLFTARGFAEVSVDDV
MSMEG_1420|M.smegmatis_MC2_155 -----MSEGSRAGRRRSTTQD------HIAGVAIDLFAARGFDAVSVDDV
MAB_3837|M.abscessus_ATCC_1997 ----MNAQPARVGRRPSTTRD------EITAVAMTLFAQHGFDEVSVDDI
MLBr_00064|M.leprae_Br4923 MINSTAGQTSQSRSRRSTRPSGDDRELAILATAEQLLEDRPLTEISVDYL
.* *: : : :*** :
Mflv_5062|M.gilvum_PYR-GCK ASAAGIARRTLFRYYPSKSALPWGDFDAHLDHMRALLADLDP--EVPIRD
Mvan_1294|M.vanbaalenii_PYR-1 AAAAGIARRTLFRYYPSKSALPWGDFDAHLEHMRELLDELDP--SVPIRD
TH_1899|M.thermoresistible__bu AAAAGIGRRTLFRYYPSKNAIPWGDFDAHLDRMRDLLDGIGP--DVPIGA
MUL_0771|M.ulcerans_Agy99 AQAAGIARRTLFRYYASKNAIPWGDFDTHLAQLQDLLERIDG--HVRLGK
MMAR_1019|M.marinum_M AQAAGIARRTLFRYYASKNAIPWGDFDTHLAQLQDLLERIDG--HVRLGK
Mb0710c|M.bovis_AF2122/97 ARAAGIARRTLFRYYASKNAIPWGDFSTHLAQLQGLLDNIDS--RIQLRD
Rv0691c|M.tuberculosis_H37Rv ARAAGIARRTLFRYYASKNAIPWGDFSTHLAQLQGLLDNIDS--RIQLRD
MAV_4481|M.avium_104 AAAAGISRRTLFRYYASKNAIPWGDFDTHLARLRELLDNVES--RVPLGD
MSMEG_1420|M.smegmatis_MC2_155 AAAAGISRRTLFRYYASKSAIPWGDFDSHLQHLQNLLRTLSS--EVSLGD
MAB_3837|M.abscessus_ATCC_1997 AAAAGIARRTVFRYYSSKNAIVWGDFDSHLGVMRALLDETAD--TVDTST
MLBr_00064|M.leprae_Br4923 AKSAGISRPTFYFYFASKEAVLLTLLDRVFNKANAALETLIKNRDNKRGN
* :***.* *.: *:.**.*: :. : . *
Mflv_5062|M.gilvum_PYR-GCK ALRTAL-LAFNDF--DDPDRHR-QRMRLILETEALQAYSMTMYAGWRAVV
Mvan_1294|M.vanbaalenii_PYR-1 ALRTAL-LAFNDF--DDADRHR-QRMRLILETEALQAYSMTMYAGWRAVV
TH_1899|M.thermoresistible__bu ALRSML-LAFNDFGEDETGRHR-RRMRVILQTDALQAHSMTMYAGWRAVV
MUL_0771|M.ulcerans_Agy99 ALREAL-LAFNTYDESETIRHR-QRMRIILQTAELQAYSMTMYAGWRAVI
MMAR_1019|M.marinum_M ALREAL-LAFNTYDESETIRHR-QRMRIILQTAELQAYSMTMYAGWRAVI
Mb0710c|M.bovis_AF2122/97 ALRAAL-LAFNTFDESETIRHR-KRMRVILQTPELQAYSMTMYAGWREVI
Rv0691c|M.tuberculosis_H37Rv ALRAAL-LAFNTFDESETIRHR-KRMRVILQTPELQAYSMTMYAGWREVI
MAV_4481|M.avium_104 ALRTAL-LAFNTFDESETARHR-QRMRVILQTAELQAYSMTMYAGWREVI
MSMEG_1420|M.smegmatis_MC2_155 ALREAL-LTFNTYADHEMAEHR-QRMRVILETEELQAYSMTMYAGWRTVI
MAB_3837|M.abscessus_ATCC_1997 ALRQAV-LTFNDFPVDETPRHR-MRMRMILEVPALQAYSMLMYTGWRDVI
MLBr_00064|M.leprae_Br4923 KWRTGINVFFETFGSHKAVTCAGQVTRDNTEVRKLWSTFMQKWISYTAAV
* : : *: : . * :. * : * : .: .:
Mflv_5062|M.gilvum_PYR-GCK AAFVADRLGVGGDDLRPQTVAWTM-LAVSLSAYEHWLADE-TACLQSSLG
Mvan_1294|M.vanbaalenii_PYR-1 AAFVAGRLGVDPAELMPQTVAWTM-LAVSLSAYEYWLADE-SVSLAAALG
TH_1899|M.thermoresistible__bu ADFVARRLGLAPSDLVPQTVAWTM-LGVALSAYAYWLADE-SVPLAQALA
MUL_0771|M.ulcerans_Agy99 AGFVARRLSVKPTDLVPQTVAWTM-LGVALSAYEHWLSDE-SVSLPEALG
MMAR_1019|M.marinum_M AGFVARRLSVKPTDLVPQTVAWTM-LGVALSAYEHWLSDE-SVSLPEALG
Mb0710c|M.bovis_AF2122/97 AKFVARRSGGKTTDFMPQTVAWTM-LGVALSAYEHWLRDE-SVSLTEALG
Rv0691c|M.tuberculosis_H37Rv AKFVARRSGGKTTDFMPQTVAWTM-LGVALSAYEHWLRDE-SVSLTEALG
MAV_4481|M.avium_104 AGFVARRCECKTTDLAPQTVAWMM-LGVALSAYEHWLGDE-SVPLPVALG
MSMEG_1420|M.smegmatis_MC2_155 AEYVADRLGMSTTDLMPQTVAWLM-LGVALSAYENWLADE-SVSLRGAIA
MAB_3837|M.abscessus_ATCC_1997 AQFVARRSGTDPGSLLPQTVAWTL-LGVALAAYEQWLADE-SSKLTTLLG
MLBr_00064|M.leprae_Br4923 IDAERKR-GVAPDTLPATELATALNLMNERALFASFTAEQPHVPESRVLD
* : . :* : * : : : :: :
Mflv_5062|M.gilvum_PYR-GCK EAFDLLASGLSGLESGVSEFAPGARR
Mvan_1294|M.vanbaalenii_PYR-1 DAFDMLASGLAGLETGVSEFGA----
TH_1899|M.thermoresistible__bu DAFDAIRPGLDALDG--KKFA-----
MUL_0771|M.ulcerans_Agy99 NAFDVVGAGLEKLD------------
MMAR_1019|M.marinum_M NAFDVVGAGLEKLD------------
Mb0710c|M.bovis_AF2122/97 AAFDVVGAGLDRLNQ-----------
Rv0691c|M.tuberculosis_H37Rv AAFDVVGAGLDRLNQ-----------
MAV_4481|M.avium_104 NAFDVVGAGLNGLDR-----------
MSMEG_1420|M.smegmatis_MC2_155 AAFEVARPGLDALQPAH---------
MAB_3837|M.abscessus_ATCC_1997 KGFDAAEVGLRALR------------
MLBr_00064|M.leprae_Br4923 TLVHIWVSSIYNEIR-----------
.. .: