For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIFVVDARAPIALWRSLRAMTEFRRLLELRAVSQFGDGLFQAGLAGAILFNVERAAGPWQIAGSFAVLFL PYSALGPFAGALLDRWDRRLVMIGANLGRLLLVVLVGVLLALGAGDLEILCCALIINGFTRFVSSGLSAA LPDVVPRGQVVAMNSVATATGAAAAFLGANFMLLPRWLFGAGDAGASVVIFIVLIPVALALWLSVRFGPH VLGPHESKRAIHGSVAYAVSTGWLHGARTVASVRTVAATLGGLAAHRAAFGINTLIVLVLVRHTDTPTQV AGVGLGTAVVFVGATGAGTFLATLVMPAVVARFGRYATANGALAFAALVQAVALSLHLPVMLVCGFLLGA AGQFVKLCADSAMQIDVDDALRGHVFTVQDALFWMSFVCAIAVAALVIAPDGHSPGLVAAGSVIYLAGLG LHAFVASPRRA
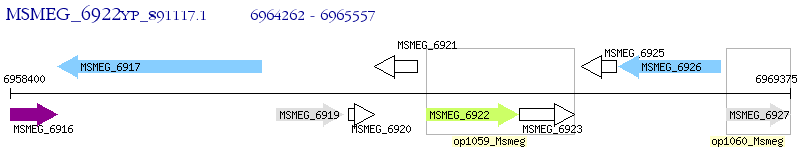
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6922 | - | - | 100% (431) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_6390 | - | 3e-09 | 22.28% (386) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_5674 | - | 5e-09 | 27.98% (386) | membrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0038c | - | 1e-157 | 66.11% (419) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_0849 | - | 1e-164 | 69.05% (420) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0037c | - | 1e-157 | 66.11% (419) | integral membrane protein |
| M. leprae Br4923 | MLBr_02143 | - | 6e-05 | 21.45% (387) | hypothetical protein MLBr_02143 |
| M. abscessus ATCC 19977 | MAB_4929 | - | 1e-130 | 57.73% (414) | hypothetical protein MAB_4929 |
| M. marinum M | MMAR_0052 | - | 1e-154 | 64.44% (419) | integral membrane protein |
| M. avium 104 | MAV_0051 | - | 1e-153 | 63.18% (421) | transporter, major facilitator family protein |
| M. thermoresistible (build 8) | TH_1066 | - | 7e-06 | 21.24% (372) | POSSIBLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_0051 | - | 1e-153 | 63.96% (419) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_6058 | - | 1e-170 | 70.85% (422) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0849|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_6058|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6922|M.smegmatis_MC2_155 --------------------------------------------------
Mb0038c|M.bovis_AF2122/97 --------------------------------------------------
Rv0037c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0052|M.marinum_M --------------------------------------------------
MUL_0051|M.ulcerans_Agy99 --------------------------------------------------
MAV_0051|M.avium_104 --------------------------------------------------
MAB_4929|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02143|M.leprae_Br4923 MSGRRRNHPGRLASIPGLRTRTGSRNQHPGIANYPADSSDFRPAQQRRAL
TH_1066|M.thermoresistible__bu ------------------------------MANYPSDDPAPRRPGRPKAP
Mflv_0849|M.gilvum_PYR-GCK ---------------------------------------MANERSPVAYL
Mvan_6058|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6922|M.smegmatis_MC2_155 --------------------------------------------------
Mb0038c|M.bovis_AF2122/97 --------------------------------------------------
Rv0037c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0052|M.marinum_M --------------------------------------------------
MUL_0051|M.ulcerans_Agy99 --------------------------------------------------
MAV_0051|M.avium_104 --------------------------------------------------
MAB_4929|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02143|M.leprae_Br4923 EQREQQAGQLDPARRSPSMPSANRYLPPLGQQQSEPQHNSAPPCGPYPGE
TH_1066|M.thermoresistible__bu PSANRWLPPLDEDR--PTFDRGAGEPPPRGYRSTG---------RPRSDE
Mflv_0849|M.gilvum_PYR-GCK YCGSGSARPHPPQDVTPVPDDRAPRTSWRAVQALPQFRRLLELRAVSQFA
Mvan_6058|M.vanbaalenii_PYR-1 -----------------MPDDRAPRTLWRSVRALPEFRRLLELRAVSQFG
MSMEG_6922|M.smegmatis_MC2_155 --------------MIFVVDARAPIALWRSLRAMTEFRRLLELRAVSQFG
Mb0038c|M.bovis_AF2122/97 -----MPRVEVGLVIHSRMHARAPVDVWRSVRSLPDFWRLLQVRVASQFG
Rv0037c|M.tuberculosis_H37Rv -----MPRVEVGLVIHSRMHARAPVDVWRSVRSLPDFWRLLQVRVASQFG
MMAR_0052|M.marinum_M ------------------MHASAPVELWRSVRSLPDFWRLLQVRIASQFG
MUL_0051|M.ulcerans_Agy99 ------------------MHASAPVELWRSVRSLPDFWRLLQVRIASQFG
MAV_0051|M.avium_104 -------------MIPTRMQSSAPVEIWRSVRALPDFWRLLQVRVASQFG
MAB_4929|M.abscessus_ATCC_1997 -----------------MTARHSPLSLWRAIHDLPDFGRLVWVRLLTQFS
MLBr_02143|M.leprae_Br4923 RIKVTRAAAQRSREMGYRMYWMVQRAATADGADKSGLTALTWPVVTNFAV
TH_1066|M.thermoresistible__bu RITVTRAAAQRSREMGSRMYGLVHRAATADGADKSGLTALTWPVVANFAV
. : * .
Mflv_0849|M.gilvum_PYR-GCK EGLFQAGIAGALLFNPEREADPWAIAGAFAALFLPYSLLGPFAGALLDRW
Mvan_6058|M.vanbaalenii_PYR-1 DGLFQAGIAGAILFNPEREAEPWAIAAAFAVLFLPYSLLGPFAGALLDRW
MSMEG_6922|M.smegmatis_MC2_155 DGLFQAGLAGAILFNVERAAGPWQIAGSFAVLFLPYSALGPFAGALLDRW
Mb0038c|M.bovis_AF2122/97 DGLFQAGLAGALLFNPDRAADPMAIAGAFAVLFLPYSLLGPFAGALMDRW
Rv0037c|M.tuberculosis_H37Rv DGLFQAGLAGALLFNPDRAADPMAIAGAFAVLFLPYSLLGPFAGALMDRW
MMAR_0052|M.marinum_M DGLFQAGLAGALLFNPDRAADPMAIARAFAVLFLPYSLLGPFAGALMDRW
MUL_0051|M.ulcerans_Agy99 DGLFQAGLAGALLFNPDRAADPMAIARAFAVLFLPYSLLGPFAGALMDRW
MAV_0051|M.avium_104 DGLFQAALAGALLFNPDRAADPLAIARAFTVLFLPYSLLGPFAGALMDRW
MAB_4929|M.abscessus_ATCC_1997 DGLFQAGLAGALLFNPEREAEPWAIAGAFAVLFLPYSAIGPFAGALLDRW
MLBr_02143|M.leprae_Br4923 DSAMAVALANTLFFAAASGESKSKVALYLLITIAPFAVIAPLIGPALDRL
TH_1066|M.thermoresistible__bu DAAMAVALANTLFFAAATGESKARVALYLLITIAPFAVIAPLIGPALDRL
:. : ..:*.:::* :* : : *:: :.*: *. :**
Mflv_0849|M.gilvum_PYR-GCK DR--RLVLIGANTGRVAIVLLVGVLLAAGAG---DIPILLSALVANGFTR
Mvan_6058|M.vanbaalenii_PYR-1 DR--RLVLIGANCGRLVVILVVGALLSAGLS---DIPILLSALLANGFTR
MSMEG_6922|M.smegmatis_MC2_155 DR--RLVMIGANLGRLLLVVLVGVLLALGAG---DLEILCCALIINGFTR
Mb0038c|M.bovis_AF2122/97 DR--RWVLVGANTGRLALIAGVGTILAVGAG---DVPLLVGALVANGLAR
Rv0037c|M.tuberculosis_H37Rv DR--RWVLVGANTGRLALIAGVGTILAVGAG---DVPLLVGALVANGLAR
MMAR_0052|M.marinum_M DR--RLVIVGANIGRLILIAGIGTILAFEAG---DLLLLFVALLANGLAR
MUL_0051|M.ulcerans_Agy99 DR--RLVIVGANIGRLILIAGIGTILAFEAG---DLLLLFVALLANGLAR
MAV_0051|M.avium_104 DR--RLVLVGANVGRLVLIAAIGTILAVRAG---DLSLLLGALFANGLAR
MAB_4929|M.abscessus_ATCC_1997 DR--RAVLICANLIRVLLVALVGFELAFSAD---DVVLLLGALIVNGVTR
MLBr_02143|M.leprae_Br4923 QHGRRVALATSFVLRTGLATLLIMNYDGATGSYPSMVLYPCALAMMVLSK
TH_1066|M.thermoresistible__bu QHGRRVALATSFGLRTALVVVLIANYDGATGSFPSWVLYPCALAMMVLSK
:: * .: : * : : . . : ** .::
Mflv_0849|M.gilvum_PYR-GCK FVSSGLSAALPHVVP-REQVIPMNSVANLISSIGAFAGAVFMLGPRWLFG
Mvan_6058|M.vanbaalenii_PYR-1 FVSSGLSAALPHVVP-RDQVIAMNSVATATTSIAAFAGAIFMLLPRWLFG
MSMEG_6922|M.smegmatis_MC2_155 FVSSGLSAALPDVVP-RGQVVAMNSVATATGAAAAFLGANFMLLPRWLFG
Mb0038c|M.bovis_AF2122/97 FVASGLSAALPHVVP-REQVVTMNSVAIASGAVSAFLGANFMLLPRWLLG
Rv0037c|M.tuberculosis_H37Rv FVASGLSAALPHVVP-REQVVTMNSVAIASGAVSAFLGANFMLLPRWLLG
MMAR_0052|M.marinum_M FVASGLSASLPHVVP-REQVVTMNSVATASGAVAAFVGANFMLLPRWLAG
MUL_0051|M.ulcerans_Agy99 FVASGLSASLPHVVP-REQVVTMNSVATASGAVAAFVGANFMLLPRWLAG
MAV_0051|M.avium_104 FVGSGLSASLPHVVP-REQVVTMNAVATAAGAVAAFLGANFMLVPRFLFG
MAB_4929|M.abscessus_ATCC_1997 FVTSGLSAALPHVVP-RDQVATMNSVATAVGATATFIGANFMLMLRAAFG
MLBr_02143|M.leprae_Br4923 SFSVLRSAVTPRVMPPSIDLVRVNSRLTVFGLLGGTIAGGAIAAGVEFVC
TH_1066|M.thermoresistible__bu SFSVLRSAMTPRVLPPTIDLVRVNSRLTMFGLLGGTMLGGAVAAAAEYVF
. ** * *:* :: :*: . . :
Mflv_0849|M.gilvum_PYR-GCK AGDSGASITILMVTVPVGLALWLSMRFPPRLLGPDDSKRAVHGS-----V
Mvan_6058|M.vanbaalenii_PYR-1 AGDSGASVIIFMVAVPVSVALWLSMRFPPRLLGPDDSARAVHGS-----V
MSMEG_6922|M.smegmatis_MC2_155 AGDAGASVVIFIVLIPVALALWLSVRFGPHVLGPHESKRAIHGS-----V
Mb0038c|M.bovis_AF2122/97 SGDEGASAIVFLVAIPVSIALLWSLRFGPRVLGPDDTERAIHGS-----A
Rv0037c|M.tuberculosis_H37Rv SGDEGASAIVFLVAIPVSIALLWSLRFGPRVLGPDDTERAIHGS-----A
MMAR_0052|M.marinum_M GGDHGAAAVIFTAMIPVAISLLLALRFAPRVLGPDDTKRAIHGS-----A
MUL_0051|M.ulcerans_Agy99 GGDHGAAAVIFTAMIPVAISLLLALRFAPRVLGPDDTKRAIHGS-----A
MAV_0051|M.avium_104 AGDRGAAAIVFLTVVPVSIALLLSWRFAPRALGPDDTWRAIHGP-----V
MAB_4929|M.abscessus_ATCC_1997 AGDLGSATVIFLVIVPTTAAAWVASGFPGDRLGPDDSVRAVHGS-----A
MLBr_02143|M.leprae_Br4923 AHLFKLPGALFVVAAITISGALLSMRIPRWVEVTAGEIPATLSYRLHRKP
TH_1066|M.thermoresistible__bu N-LFELPGALYVIVAVSLAGAVLSMRIPRWVEVTEGEVPATLTY---HGD
. : . : : . *
Mflv_0849|M.gilvum_PYR-GCK MYAVATGWLYGARTVLAVPTAASTLAGLAAHRMVFGINTLLMLVIVRHTE
Mvan_6058|M.vanbaalenii_PYR-1 LYAVATGWLHGARTVLAVPSVSSTLSGLAAHRMVFGVNTLLVLVMVRHTE
MSMEG_6922|M.smegmatis_MC2_155 AYAVSTGWLHGARTVASVRTVAATLGGLAAHRAAFGINTLIVLVLVRHTD
Mb0038c|M.bovis_AF2122/97 VYAVVTGWLHGARTVVQLPTVAAGLSGLAAHRMVVGINSLLILLLVRHVT
Rv0037c|M.tuberculosis_H37Rv VYAVVTGWLHGARTVVQLPTVAAGLSGLAAHRMVVGINSLLILLLVRHVT
MMAR_0052|M.marinum_M VYAVITGWMHGVRTVVQLPTVAAALSGLAAHRMVVGVNSLLILLLVHHMR
MUL_0051|M.ulcerans_Agy99 VYAVITGWMHGVRTVVQIPTVAAALSGLAAHRMVVGVNSLLILLLVHHMR
MAV_0051|M.avium_104 LYAVITGWLHGARTVAQRPTVAATLSGLAAHRMVVGINSLLVLLLVHHLP
MAB_4929|M.abscessus_ATCC_1997 FYAVATGWLHGARTIVGTPSVFDALIGLAAHRMVFGVNTLLVLVLVRSNE
MLBr_02143|M.leprae_Br4923 LRQSWPEEVKKVSGRLRQPLGRNIITSLWGNCTIKVMVGFLFLYPAFVAK
TH_1066|M.thermoresistible__bu LRR--HPDQRRHSGAARQPMGRNTITALWGNCTIKLMVGFLFLYPAFVAK
: .* .: : ::.* .
Mflv_0849|M.gilvum_PYR-GCK TATVAG---FGTTVLFVIAGGAGQFVATIATPALVTRWGRYAAPNGALAL
Mvan_6058|M.vanbaalenii_PYR-1 AATVAG---FGTTVLFVSAGGAGQFLATVVTPTLVKKWGRYATPNGALAF
MSMEG_6922|M.smegmatis_MC2_155 TPTQVAGVGLGTAVVFVGATGAGTFLATLVMPAVVARFGRYATANGALAF
Mb0038c|M.bovis_AF2122/97 ARAVGG---LGTALLFFAATGLGAFLANVLTPTAIRRWGRYATANGALAA
Rv0037c|M.tuberculosis_H37Rv ARAVGG---LGTALLFFAATGLGAFLANVLTPTAIRRWGRYATANGALAA
MMAR_0052|M.marinum_M DVDVEG---LGTALLFFAATGLGAFLANVLTPGLIRRWGRYATTNGALAA
MUL_0051|M.ulcerans_Agy99 DVDVEG---LGTALLFFAATGLGAFLANVLTPGLIRRWGRYATTNGALAA
MAV_0051|M.avium_104 GLEGGG---FGTALLFFGAAGLGAFLANVLTPPAIRRWGRYATANGALAA
MAB_4929|M.abscessus_ATCC_1997 S-VFAG---IGAAALFLACTGIGSFLGTVSTPALLRRFGRGRTLGMALGG
MLBr_02143|M.leprae_Br4923 EHQANGWVQLGMLGLIGTAAAIGNFAGNFTSARLQLGRPAVLVVRCTIAV
TH_1066|M.thermoresistible__bu SHEATGLEQLMILGLIGAAAAVGNFIGNFTAARLELGHPAQLVVRAAIAV
. : :: . . * * ... . . ::.
Mflv_0849|M.gilvum_PYR-GCK AAVVQVCAATLQLPVMIVCGFVLGAAGQVVKLCADSAMQIDVDDALRGHM
Mvan_6058|M.vanbaalenii_PYR-1 AAAVQLCGATLNLPVMIVCGFLLGAAGQVVKLCADTAMQIDVEDALRGHL
MSMEG_6922|M.smegmatis_MC2_155 AALVQAVALSLHLPVMLVCGFLLGAAGQFVKLCADSAMQIDVDDALRGHV
Mb0038c|M.bovis_AF2122/97 AATIQVAAAGLLVPVMVVCGFLLGVAGQVVKLCADSAMQMDVDDALRGHV
Rv0037c|M.tuberculosis_H37Rv AATIQVAAAGLLVPVMVVCGFLLGVAGQVVKLCADSAMQMDVDDALRGHV
MMAR_0052|M.marinum_M AAVIQLAGAGLLLPVMVVCGFLLGVAGQVVKLCADSAMQIDVDDALRGHV
MUL_0051|M.ulcerans_Agy99 AAVTQLAGAGLLLPVMVVCGFLLGVAGQVVKLCADSAMQIDVDGALRGHV
MAV_0051|M.avium_104 SAIIEVAGAELLLPVMVVCGFLLGVTGQMVKLCADSAMQMDVDDALRGHV
MAB_4929|M.abscessus_ATCC_1997 AVVLQLIASGLYIPILLASAFGLGMAGQVIKLCADVGMQTDVDDALRGHV
MLBr_02143|M.leprae_Br4923 TVLALAASVAGNLLMTTIATLITSGSSAIAKASLDASLQNDLPEESRASG
TH_1066|M.thermoresistible__bu TIGALATAVTGELLVAALATLITSGASAIAKASLDASLQDDLPEQSRASA
: . : : . : . :. . * . * .:* *: *.
Mflv_0849|M.gilvum_PYR-GCK FAVQDSLFWVAFIVAITGAAAVIPADGHSPALAVAGTGLYLAGLAAHAV-
Mvan_6058|M.vanbaalenii_PYR-1 FAVQDALFWLAFIVAISAAAFVIPPDGRSVGLAVAGVGTYLAGLAVHAT-
MSMEG_6922|M.smegmatis_MC2_155 FTVQDALFWMSFVCAIAVAALVIAPDGHSPGLVAAGSVIYLAGLGLHAF-
Mb0038c|M.bovis_AF2122/97 FAVQDALFWVSYILSITVAAALIPEHGHAPVFVLFGSAIYLAGLVVHTI-
Rv0037c|M.tuberculosis_H37Rv FAVQDALFWVSYILSITVAAALIPEHGHAPVFVLFGSAIYLAGLVVHTI-
MMAR_0052|M.marinum_M FAVQDALFWVSYIVSVTVAAALIPDDGRAPVFVLVGSVIYLIGLCVHSI-
MUL_0051|M.ulcerans_Agy99 FAVQDALFWVSYIVSVTVAAALIPDDGRAPVFVLVGSLIYLLGLCVHSI-
MAV_0051|M.avium_104 FAVQDALFWVAFIVAITVAGMVIPDDGHAPVFALFGSVLYLVGLAVHGI-
MAB_4929|M.abscessus_ATCC_1997 FAVQDSLFWVTFTGAMAVGAAFIT---HAHWLAFSGAMIYLAGLIVHMTR
MLBr_02143|M.leprae_Br4923 FGRSESTLQLAWVLGGALGVMVYTDLWVGFTAVSALLILGLAQTVVSFRG
TH_1066|M.thermoresistible__bu FGRSESLLQLAWVAGGATGVLVYTELWIGFTTISAILILGLAQTIVSYRG
* .:: : ::: . : . . . . *
Mflv_0849|M.gilvum_PYR-GCK -----------------MGRRGR-RG-------------
Mvan_6058|M.vanbaalenii_PYR-1 -----------------VGRRGRPRG-------------
MSMEG_6922|M.smegmatis_MC2_155 -----------------VASPRRA---------------
Mb0038c|M.bovis_AF2122/97 -----------------VGRR--GQPVIGR---------
Rv0037c|M.tuberculosis_H37Rv -----------------VGRR--GQPVIGR---------
MMAR_0052|M.marinum_M -----------------VGRRSSAANVEGG---------
MUL_0051|M.ulcerans_Agy99 -----------------VGRRSSASNVEGC---------
MAV_0051|M.avium_104 -----------------VGRR--GE--------------
MAB_4929|M.abscessus_ATCC_1997 TLWPSTLDTGSLPTQPNAGSLPTQPNPEGQARDLDR---
MLBr_02143|M.leprae_Br4923 DSLIPG----------LGGNRPVMIEQESMRRAAAVSPQ
TH_1066|M.thermoresistible__bu DSLIPG----------FGGNRPVLAEPEGRNDHPAFTPR
.