For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVTYRSVEVSQPGDALRITERTVREPGPGEVRIRVLACGVCHSDSQIAAGHLPGTVFPVTPGHEVAGQI DAIGAGVEEWQIGDRVAVGWFGFYCGSCYRCRRGDFVHCERSKVTGAAFPGGYAEMLVVHQSGLARIPDT LSATDAAPLACAGVTMFHSLRESGARPGDVVAILGVGGLGHLGVQFAAKMGFVTVAIARGNEKADMAMQL GAHHYIDATADDVAKKLRALGGARVVAATATNPAAISATVDGLAPHGELLTLAVLGESLHVTPLQLINSS GTVHGHPAGVAADIEDTLEFAAVHGIRPWIEPMPLHEAATGYDRMMRNEARFRVVLTMPQPA
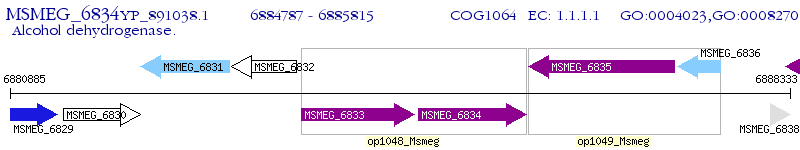
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6834 | - | - | 100% (342) | alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3464 | - | 1e-95 | 51.34% (335) | alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6833 | - | 1e-86 | 48.50% (334) | alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3071 | adhC | 4e-47 | 32.65% (340) | NADP-dependent alcohol dehydrogenase ADHC |
| M. gilvum PYR-GCK | Mflv_0236 | - | 1e-101 | 52.54% (335) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3045 | adhC | 4e-47 | 32.65% (340) | NADP-dependent alcohol dehydrogenase ADHC |
| M. leprae Br4923 | MLBr_01730 | adhA | 5e-48 | 30.70% (342) | alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3400 | - | 2e-44 | 31.29% (342) | NADP-dependent alcohol dehydrogenase C |
| M. marinum M | MMAR_2782 | adhC | 1e-104 | 53.73% (335) | NADP-dependent alcohol dehydrogenase AdhC |
| M. avium 104 | MAV_2812 | - | 1e-100 | 55.59% (322) | aldehyde dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_4581 | - | 2e-47 | 32.65% (340) | PUTATIVE alcohol dehydrogenase, zinc-containing |
| M. ulcerans Agy99 | MUL_2965 | adhC | 1e-104 | 53.73% (335) | NADP-dependent alcohol dehydrogenase AdhC |
| M. vanbaalenii PYR-1 | Mvan_0669 | - | 1e-100 | 53.13% (335) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2782|M.marinum_M ----------------MATHKAVQIGAAGGLLQLVDVDTAPPGDGEVRIS
MUL_2965|M.ulcerans_Agy99 ----------------MATHKAVQIGAAGGPLQLVDVDTAPPADGEVRIS
MAV_2812|M.avium_104 -------------------------------MELVDTETRPPGRDEVRLT
Mflv_0236|M.gilvum_PYR-GCK ----------------MPTHKAVHVAGANEDLTLVEVETAPPPPGHVRLA
Mvan_0669|M.vanbaalenii_PYR-1 ----------------MPTHKAVQVPSANEALTHVEVDTLPPPAGHVRID
MSMEG_6834|M.smegmatis_MC2_155 ---------------MTVTYRSVEVSQPGDALRITERTVREPGPGEVRIR
Mb3071|M.bovis_AF2122/97 ----------------MSTVAAYAAMSATEPLTKTTITRRDPGPHDVAID
Rv3045|M.tuberculosis_H37Rv ----------------MSTVAAYAAMSATEPLTKTTITRRDPGPHDVAID
MLBr_01730|M.leprae_Br4923 MKSHVSKHRRKRRLGAMTTVSAYAATSATEPLTKVTINRRAPGPHDVAIQ
TH_4581|M.thermoresistible__bu ----------------MTTVSAYAATSVSEPLTRTTITRREMGPHDVTIA
MAB_3400|M.abscessus_ATCC_1997 ----------------MVTVSAYAATSGKDPLVPTTIERREVGPHDVFID
: . .* :
MMAR_2782|M.marinum_M VSACGICGTDRAFVHGAFPGLGWPLTPGHEIAGRIAEIGNGVEDFAVGDR
MUL_2965|M.ulcerans_Agy99 VSACGICGTDRAFVHGAFPGLDWPLTPGHEIAGRIAEIGNGVEDFAVGDR
MAV_2812|M.avium_104 VGACGICGTDAHFVNGAFPGLSWPLTPGHEIAGRIAELGEGVEGFAVGQR
Mflv_0236|M.gilvum_PYR-GCK VKACGVCGTDREFRAGHFPGLQWPLTLGHEIAGAVAEVGDGVEDFSVGDR
Mvan_0669|M.vanbaalenii_PYR-1 VAACGVCGTDREFRAGHFPGLQWPITLGHEIAGTVAELGDGVEEFAVGDR
MSMEG_6834|M.smegmatis_MC2_155 VLACGVCHSDSQIAAGHLPGTVFPVTPGHEVAGQIDAIGAGVEEWQIGDR
Mb3071|M.bovis_AF2122/97 IKFAGICHSDIHTVKAEWGQPNYPVVPGHEIAGVVTAVGSEVTKYRQGDR
Rv3045|M.tuberculosis_H37Rv IKFAGICHSDIHTVKAEWGQPNYPVVPGHEIAGVVTAVGSEVTKYRQGDR
MLBr_01730|M.leprae_Br4923 IKFAGICHSDIHTVKAEWGQPNYPVVPGHEIAGVVTAVGTEVTKHKVGDR
TH_4581|M.thermoresistible__bu IHFAGICHSDIHTVKGEWGEPAFPLVPGHEIAGVVTAVGSEVTRYRVGDR
MAB_3400|M.abscessus_ATCC_1997 IKFAGICHSDIHTVRDEWGRANYPIVPGHEIAGVITEVGSEVTKFKPGDH
: .*:* :* :*:. ***:** : :* * *::
MMAR_2782|M.marinum_M VAVGWFGGNCNHCDPCRKGMFIHCAK--------MKVPGWQYPGGYAESV
MUL_2965|M.ulcerans_Agy99 VAVGWFGGNCNHCDPCRKGMFIHCSK--------MKVPGWQYPGGYAESV
MAV_2812|M.avium_104 VAVGWFGGCCYRCDPCRAGLFIHCVN--------GKVPSWHYPGGYAESV
Mflv_0236|M.gilvum_PYR-GCK VAVGWFGGNCNRCIPCRRGSFMQCER--------MQVPSWHYPGGYAESV
Mvan_0669|M.vanbaalenii_PYR-1 VAVGWFGGNCNRCVPCRRGFFMQCER--------MQVPSWQYPGGYAESV
MSMEG_6834|M.smegmatis_MC2_155 VAVGWFGFYCGSCYRCRRGDFVHCER--------SKVTGAAFPGGYAEML
Mb3071|M.bovis_AF2122/97 VGVGCFVDSCRECNSCTRGIEQYCKPGANFTYNSIGKDGQPTQGGYSEAI
Rv3045|M.tuberculosis_H37Rv VGVGCFVDSCRECNSCTRGIEQYCKPGANFTYNSIGKDGQPTQGGYSEAI
MLBr_01730|M.leprae_Br4923 VGVGCMVGSCGECSSCLAGIEQYCKRGATFTYNSIGTDGQSTQGGYSQAI
TH_4581|M.thermoresistible__bu VGVGCFVDSCRECRHCLAGDEQYCVRGMTGTYNGVDRDGRPTQGGYSGAI
MAB_3400|M.abscessus_ATCC_1997 AGVGCFVNSCRECPSCLAGLEQFCERGMVGTYGAKDRDGTQTQGGYSQAI
..** : * * * * * . ***: :
MMAR_2782|M.marinum_M TVPATALARIPAELSDAEAAPMGCAGVTTYNALRHTRARPGDRVAVLGVG
MUL_2965|M.ulcerans_Agy99 TVPATALARIPAELSDAEAAPMGCAGVTTYNALRHTRARPGDRVAVLGVG
MAV_2812|M.avium_104 TVPTSALARIPEELTDAEAAPMGCAGVTTYNALRHTKALPGDRVAILGVG
Mflv_0236|M.gilvum_PYR-GCK TVPWTALARIPDGLSFVEAAPMGCAGVTTFNGLRQTRAKAGDVVAVLGVG
Mvan_0669|M.vanbaalenii_PYR-1 TAPATALARIPDGLSFVEAAPMGCAGVTTFNGLRRTRAKAGDLVAVLGVG
MSMEG_6834|M.smegmatis_MC2_155 VVHQSGLARIPDTLSATDAAPLACAGVTMFHSLRESGARPGDVVAILGVG
Mb3071|M.bovis_AF2122/97 VVDENYVLRIPDVLPLDVAAPLLCAGITLYSPLRHWNAGANTRVAIIGLG
Rv3045|M.tuberculosis_H37Rv VVDENYVLRIPDVLPLDVAAPLLCAGITLYSPLRHWNAGANTRVAIIGLG
MLBr_01730|M.leprae_Br4923 VVDENFVVHIPDALPLDAAAPLLCAGITLYSPLRHWNAKPGTRLAIIGLG
TH_4581|M.thermoresistible__bu VVDENYVLRIPDSLPLDAAAPLLCAGITTYSPLRHWNVGPGTRVAVVGLG
MAB_3400|M.abscessus_ATCC_1997 VVDENYVLRIPDSLPLDAAAPLLCAGITTFSPLRHWNAGPGKRVAVVGLG
.. . : :** *. ***: ***:* : **. . .. :*::*:*
MMAR_2782|M.marinum_M GLGHLGLQFCRAMGFETVAIARGSAKAEDARQLGAHHYIDSTTGDVAEAL
MUL_2965|M.ulcerans_Agy99 GLGHLGLQFCRAMGFETVAIARGSAKAEDARQLGAHHYIDSTTGDVAEGL
MAV_2812|M.avium_104 GLGHLGVQFAAAMGFETIAINRGTGKADDARKLGAHHYIDSTAEDPAEAL
Mflv_0236|M.gilvum_PYR-GCK GLGHLGVQWSRAMGFETVAIARGTGKAEEARELGAHHYIDSTGQDVAAEL
Mvan_0669|M.vanbaalenii_PYR-1 GLGHLGVQWARAMGFETVAIGRGTGKAEEATQFGAHHYIDSTGQDVAAEL
MSMEG_6834|M.smegmatis_MC2_155 GLGHLGVQFAAKMGFVTVAIARGNEKADMAMQLGAHHYIDATADDVAKKL
Mb3071|M.bovis_AF2122/97 GLGHMGVKLGAAMGADVTVLSQSLKKMEDGLRLGAKSYYATADPDTFRKL
Rv3045|M.tuberculosis_H37Rv GLGHMGVKLGAAMGADVTVLSQSLKKMEDGLRLGAKSYYATADPDTFRKL
MLBr_01730|M.leprae_Br4923 GLGHMGVKLGRKMGCEVTVLSQSLKKMEDGLQLGASNYYATADPDTFRKL
TH_4581|M.thermoresistible__bu GLGHMAVKLGKALGADVTVLSQSLKKREDGLRLGAGAYYATSDPDTFRAL
MAB_3400|M.abscessus_ATCC_1997 GLGHMAVKLGKAMGAEVTVLSQSLKKMEDGLRLGASHYYATADPDTFKSL
****:.:: :* . .: :. * : . .:** * :: * *
MMAR_2782|M.marinum_M RRIGGAAVVLATAGNSAAMAETVGGLLPRGELVVIGVTADPLPISPVQLI
MUL_2965|M.ulcerans_Agy99 RRIGGAAVVLATAGNSAAMAETVGGLLPRGELVVIGVTADPLPISPVQLI
MAV_2812|M.avium_104 RALGGAAVVLATAGNSQAMAATVGGLAPQGELVTIGVTPEPLPISPLQLI
Mflv_0236|M.gilvum_PYR-GCK KKLGGAAVVLATANNAEAMAATVGGLGPAGELVVVGVSADNLPISPLHLI
Mvan_0669|M.vanbaalenii_PYR-1 TKLGGASVVLATANNAAAMAAAVGGLGPAGELVVVGVTAENLPISPLDLI
MSMEG_6834|M.smegmatis_MC2_155 RALGGARVVAATATNPAAISATVDGLAPHGELLTLAVLGESLHVTPLQLI
Mb3071|M.bovis_AF2122/97 R--GGFDLILNTVSANLDLGQYLNLLDVDGTLVELGIPEHPMAVPAFALA
Rv3045|M.tuberculosis_H37Rv R--GGFDLILNTVSANLDLGQYLNLLDVDGTLVELGIPEHPMAVPAFALA
MLBr_01730|M.leprae_Br4923 R--GSFDLILNTVSANLDLGKYLNLLDVNGTLVELGIPEHPMLVPAFALA
TH_4581|M.thermoresistible__bu R--SSFDVVINTVSANLDLGRYLMLLDVDGTLVELGMPEKPMTVPAAPLV
MAB_3400|M.abscessus_ATCC_1997 R--NSFDLILNTVSSNLDMGKYLGLLGVDGTLVELGLPENPIEVPGFPLL
.. :: *. :. : * * *: :.: . : :. *
MMAR_2782|M.marinum_M NPGLSVTGHPSGTAKDVEETMHFAVLSGVRAWIEERPLAQAADGYEAMEQ
MUL_2965|M.ulcerans_Agy99 NPGLSVTGHPSGTAKDVEDTMHFAVLSGVRAWIEERPLAQAADGYEAMEQ
MAV_2812|M.avium_104 TPGLSIVGHPSGTAKDVEDTMQFAVLSGVRAWIEELPLSRAADGYAALEG
Mflv_0236|M.gilvum_PYR-GCK NDGLSVTGHPSGTSRDVEETLHFAMLSGVRARVEEVPLAQPAEAYEAMDS
Mvan_0669|M.vanbaalenii_PYR-1 NAGRSVTGHPSGTSRDVEETMHFALLSGVRAKVEEVPLARAAEAYESMDS
MSMEG_6834|M.smegmatis_MC2_155 NSSGTVHGHPAGVAADIEDTLEFAAVHGIRPWIEPMPLHEAATGYDRMMR
Mb3071|M.bovis_AF2122/97 LMRRSLAGSNIGGIAETQEMLNFCAEHGVTPEIELIEPDYINDAYERVLA
Rv3045|M.tuberculosis_H37Rv LMRRSLAGSNIGGIAETQEMLNFCAEHGVTPEIELIEPDYINDAYERVLA
MLBr_01730|M.leprae_Br4923 LMRRSLAGSNIGGITETQEMMNFSAEHSVTPEIELIEPDYINEAYERVLA
TH_4581|M.thermoresistible__bu GGRRRLAGSLIGGIAETQEMLDFCAEHGVLPEIEVIEPEYINTAYERVLA
MAB_3400|M.abscessus_ATCC_1997 TNRRSISGSLIGGIPETQEMLDFCAKHGIGSEIEVIDASYINEAYERVVA
: * * : :: :.*. .: . :* .* :
MMAR_2782|M.marinum_M GRTRYRTVLAM-----
MUL_2965|M.ulcerans_Agy99 GRTRYRTVLAM-----
MAV_2812|M.avium_104 GRAHYRTVLTM-----
Mflv_0236|M.gilvum_PYR-GCK SRARYRMVLTV-----
Mvan_0669|M.vanbaalenii_PYR-1 GRARYRMVLTM-----
MSMEG_6834|M.smegmatis_MC2_155 NEARFRVVLTMPQPA-
Mb3071|M.bovis_AF2122/97 SDVRYRFVIDISAL--
Rv3045|M.tuberculosis_H37Rv SDVRYRFVIDISAL--
MLBr_01730|M.leprae_Br4923 SDVRYRFVINISSL--
TH_4581|M.thermoresistible__bu SDVRYRFVIDTASLRG
MAB_3400|M.abscessus_ATCC_1997 SDVRYRFVIDTQTL--
. .::* *: