For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LELVDTETRPPGRDEVRLTVGACGICGTDAHFVNGAFPGLSWPLTPGHEIAGRIAELGEGVEGFAVGQRV AVGWFGGCCYRCDPCRAGLFIHCVNGKVPSWHYPGGYAESVTVPTSALARIPEELTDAEAAPMGCAGVTT YNALRHTKALPGDRVAILGVGGLGHLGVQFAAAMGFETIAINRGTGKADDARKLGAHHYIDSTAEDPAEA LRALGGAAVVLATAGNSQAMAATVGGLAPQGELVTIGVTPEPLPISPLQLITPGLSIVGHPSGTAKDVED TMQFAVLSGVRAWIEELPLSRAADGYAALEGGRAHYRTVLTM
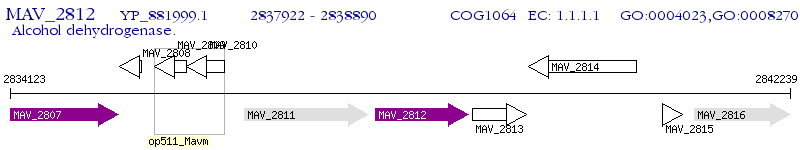
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2812 | - | - | 100% (322) | aldehyde dehydrogenase family protein |
| M. avium 104 | MAV_3911 | - | 5e-45 | 31.68% (322) | NADP-dependent alcohol dehydrogenase c |
| M. avium 104 | MAV_2857 | - | 2e-37 | 31.68% (322) | zinc-binding alcohol dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3071 | adhC | 2e-43 | 31.87% (320) | NADP-dependent alcohol dehydrogenase ADHC |
| M. gilvum PYR-GCK | Mflv_0236 | - | 1e-128 | 67.70% (322) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3045 | adhC | 2e-43 | 31.87% (320) | NADP-dependent alcohol dehydrogenase ADHC |
| M. leprae Br4923 | MLBr_01730 | adhA | 7e-44 | 30.91% (330) | alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3400 | - | 8e-48 | 33.23% (322) | NADP-dependent alcohol dehydrogenase C |
| M. marinum M | MMAR_2782 | adhC | 1e-150 | 78.26% (322) | NADP-dependent alcohol dehydrogenase AdhC |
| M. smegmatis MC2 155 | MSMEG_3464 | - | 1e-131 | 70.50% (322) | alcohol dehydrogenase |
| M. thermoresistible (build 8) | TH_4581 | - | 3e-42 | 31.56% (320) | PUTATIVE alcohol dehydrogenase, zinc-containing |
| M. ulcerans Agy99 | MUL_2965 | adhC | 1e-149 | 77.95% (322) | NADP-dependent alcohol dehydrogenase AdhC |
| M. vanbaalenii PYR-1 | Mvan_0669 | - | 1e-126 | 67.40% (319) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2782|M.marinum_M ----------------MATHKAVQIGAAGGLLQLVDVDTAPPGDGEVRIS
MUL_2965|M.ulcerans_Agy99 ----------------MATHKAVQIGAAGGPLQLVDVDTAPPADGEVRIS
MAV_2812|M.avium_104 -------------------------------MELVDTETRPPGRDEVRLT
Mflv_0236|M.gilvum_PYR-GCK ----------------MPTHKAVHVAGANEDLTLVEVETAPPPPGHVRLA
Mvan_0669|M.vanbaalenii_PYR-1 ----------------MPTHKAVQVPSANEALTHVEVDTLPPPAGHVRID
MSMEG_3464|M.smegmatis_MC2_155 ----------------MPTHRAVHVKSTGGTLELTDVETPPPDRGQVRID
Mb3071|M.bovis_AF2122/97 ----------------MSTVAAYAAMSATEPLTKTTITRRDPGPHDVAID
Rv3045|M.tuberculosis_H37Rv ----------------MSTVAAYAAMSATEPLTKTTITRRDPGPHDVAID
MLBr_01730|M.leprae_Br4923 MKSHVSKHRRKRRLGAMTTVSAYAATSATEPLTKVTINRRAPGPHDVAIQ
TH_4581|M.thermoresistible__bu ----------------MTTVSAYAATSVSEPLTRTTITRREMGPHDVTIA
MAB_3400|M.abscessus_ATCC_1997 ----------------MVTVSAYAATSGKDPLVPTTIERREVGPHDVFID
: . .* :
MMAR_2782|M.marinum_M VSACGICGTDRAFVHGAFPGLGWPLTPGHEIAGRIAEIGNGVEDFAVGDR
MUL_2965|M.ulcerans_Agy99 VSACGICGTDRAFVHGAFPGLDWPLTPGHEIAGRIAEIGNGVEDFAVGDR
MAV_2812|M.avium_104 VGACGICGTDAHFVNGAFPGLSWPLTPGHEIAGRIAELGEGVEGFAVGQR
Mflv_0236|M.gilvum_PYR-GCK VKACGVCGTDREFRAGHFPGLQWPLTLGHEIAGAVAEVGDGVEDFSVGDR
Mvan_0669|M.vanbaalenii_PYR-1 VAACGVCGTDREFRAGHFPGLQWPITLGHEIAGTVAELGDGVEEFAVGDR
MSMEG_3464|M.smegmatis_MC2_155 VAACGVCGTDRGIVNGVFPVASWPVTPGHEIAGTVAEIGPGVEDFAVGDR
Mb3071|M.bovis_AF2122/97 IKFAGICHSDIHTVKAEWGQPNYPVVPGHEIAGVVTAVGSEVTKYRQGDR
Rv3045|M.tuberculosis_H37Rv IKFAGICHSDIHTVKAEWGQPNYPVVPGHEIAGVVTAVGSEVTKYRQGDR
MLBr_01730|M.leprae_Br4923 IKFAGICHSDIHTVKAEWGQPNYPVVPGHEIAGVVTAVGTEVTKHKVGDR
TH_4581|M.thermoresistible__bu IHFAGICHSDIHTVKGEWGEPAFPLVPGHEIAGVVTAVGSEVTRYRVGDR
MAB_3400|M.abscessus_ATCC_1997 IKFAGICHSDIHTVRDEWGRANYPIVPGHEIAGVITEVGSEVTKFKPGDH
: .*:* :* : :*:. ****** :: :* * . *::
MMAR_2782|M.marinum_M VAVGWFGGNCNHCDPCRKGMFIHCAKMKVPGWQYP--------GGYAESV
MUL_2965|M.ulcerans_Agy99 VAVGWFGGNCNHCDPCRKGMFIHCSKMKVPGWQYP--------GGYAESV
MAV_2812|M.avium_104 VAVGWFGGCCYRCDPCRAGLFIHCVNGKVPSWHYP--------GGYAESV
Mflv_0236|M.gilvum_PYR-GCK VAVGWFGGNCNRCIPCRRGSFMQCERMQVPSWHYP--------GGYAESV
Mvan_0669|M.vanbaalenii_PYR-1 VAVGWFGGNCNRCVPCRRGFFMQCERMQVPSWQYP--------GGYAESV
MSMEG_3464|M.smegmatis_MC2_155 VAIGWFGGHCGTCVPCRKGLFIHCVNGQVPSLSYP--------GGYAESV
Mb3071|M.bovis_AF2122/97 VGVGCFVDSCRECNSCTRGIEQYCKPGANFTYNSIGKDGQPTQGGYSEAI
Rv3045|M.tuberculosis_H37Rv VGVGCFVDSCRECNSCTRGIEQYCKPGANFTYNSIGKDGQPTQGGYSEAI
MLBr_01730|M.leprae_Br4923 VGVGCMVGSCGECSSCLAGIEQYCKRGATFTYNSIGTDGQSTQGGYSQAI
TH_4581|M.thermoresistible__bu VGVGCFVDSCRECRHCLAGDEQYCVRGMTGTYNGVDRDGRPTQGGYSGAI
MAB_3400|M.abscessus_ATCC_1997 AGVGCFVNSCRECPSCLAGLEQFCERGMVGTYGAKDRDGTQTQGGYSQAI
..:* : . * * * * * ***: ::
MMAR_2782|M.marinum_M TVPATALARIPAELSDAEAAPMGCAGVTTYNALRHTRARPGDRVAVLGVG
MUL_2965|M.ulcerans_Agy99 TVPATALARIPAELSDAEAAPMGCAGVTTYNALRHTRARPGDRVAVLGVG
MAV_2812|M.avium_104 TVPTSALARIPEELTDAEAAPMGCAGVTTYNALRHTKALPGDRVAILGVG
Mflv_0236|M.gilvum_PYR-GCK TVPWTALARIPDGLSFVEAAPMGCAGVTTFNGLRQTRAKAGDVVAVLGVG
Mvan_0669|M.vanbaalenii_PYR-1 TAPATALARIPDGLSFVEAAPMGCAGVTTFNGLRRTRAKAGDLVAVLGVG
MSMEG_3464|M.smegmatis_MC2_155 TAPANALARIPDELSFVEAAPMGCAGVTTYNALRHTRAVAGDVVAIIGLG
Mb3071|M.bovis_AF2122/97 VVDENYVLRIPDVLPLDVAAPLLCAGITLYSPLRHWNAGANTRVAIIGLG
Rv3045|M.tuberculosis_H37Rv VVDENYVLRIPDVLPLDVAAPLLCAGITLYSPLRHWNAGANTRVAIIGLG
MLBr_01730|M.leprae_Br4923 VVDENFVVHIPDALPLDAAAPLLCAGITLYSPLRHWNAKPGTRLAIIGLG
TH_4581|M.thermoresistible__bu VVDENYVLRIPDSLPLDAAAPLLCAGITTYSPLRHWNVGPGTRVAVVGLG
MAB_3400|M.abscessus_ATCC_1997 VVDENYVLRIPDSLPLDAAAPLLCAGITTFSPLRHWNAGPGKRVAVVGLG
.. . : :** *. ***: ***:* :. **: .. .. :*::*:*
MMAR_2782|M.marinum_M GLGHLGLQFCRAMGFETVAIARGSA-KAEDARQLGAHHYIDSTTGDVAEA
MUL_2965|M.ulcerans_Agy99 GLGHLGLQFCRAMGFETVAIARGSA-KAEDARQLGAHHYIDSTTGDVAEG
MAV_2812|M.avium_104 GLGHLGVQFAAAMGFETIAINRGTG-KADDARKLGAHHYIDSTAEDPAEA
Mflv_0236|M.gilvum_PYR-GCK GLGHLGVQWSRAMGFETVAIARGTG-KAEEARELGAHHYIDSTGQDVAAE
Mvan_0669|M.vanbaalenii_PYR-1 GLGHLGVQWARAMGFETVAIGRGTG-KAEEATQFGAHHYIDSTGQDVAAE
MSMEG_3464|M.smegmatis_MC2_155 GLGHLGVQFAAAMGFDTVVIARGRGDREQDARALGARHYIDSTSEDVAAA
Mb3071|M.bovis_AF2122/97 GLGHMGVKLGAAMGADVTVLSQSLK-KMEDGLRLGAKSYYATADPDTFRK
Rv3045|M.tuberculosis_H37Rv GLGHMGVKLGAAMGADVTVLSQSLK-KMEDGLRLGAKSYYATADPDTFRK
MLBr_01730|M.leprae_Br4923 GLGHMGVKLGRKMGCEVTVLSQSLK-KMEDGLQLGASNYYATADPDTFRK
TH_4581|M.thermoresistible__bu GLGHMAVKLGKALGADVTVLSQSLK-KREDGLRLGAGAYYATSDPDTFRA
MAB_3400|M.abscessus_ATCC_1997 GLGHMAVKLGKAMGAEVTVLSQSLK-KMEDGLRLGASHYYATADPDTFKS
****:.:: :* :. .: :. : ::. :** * :: *
MMAR_2782|M.marinum_M LRRIGGAAVVLATAGNSAAMAETVGGLLPRGELVVIGVTADPLPISPVQL
MUL_2965|M.ulcerans_Agy99 LRRIGGAAVVLATAGNSAAMAETVGGLLPRGELVVIGVTADPLPISPVQL
MAV_2812|M.avium_104 LRALGGAAVVLATAGNSQAMAATVGGLAPQGELVTIGVTPEPLPISPLQL
Mflv_0236|M.gilvum_PYR-GCK LKKLGGAAVVLATANNAEAMAATVGGLGPAGELVVVGVSADNLPISPLHL
Mvan_0669|M.vanbaalenii_PYR-1 LTKLGGASVVLATANNAAAMAAAVGGLGPAGELVVVGVTAENLPISPLDL
MSMEG_3464|M.smegmatis_MC2_155 LQDLGGAAVVLGTAGNSQAMADTIGGLAPRGELVAVGVSAEPLGVSPAQL
Mb3071|M.bovis_AF2122/97 LR--GGFDLILNTVSANLDLGQYLNLLDVDGTLVELGIPEHPMAVPAFAL
Rv3045|M.tuberculosis_H37Rv LR--GGFDLILNTVSANLDLGQYLNLLDVDGTLVELGIPEHPMAVPAFAL
MLBr_01730|M.leprae_Br4923 LR--GSFDLILNTVSANLDLGKYLNLLDVNGTLVELGIPEHPMLVPAFAL
TH_4581|M.thermoresistible__bu LR--SSFDVVINTVSANLDLGRYLMLLDVDGTLVELGMPEKPMTVPAAPL
MAB_3400|M.abscessus_ATCC_1997 LR--NSFDLILNTVSSNLDMGKYLGLLGVDGTLVELGLPENPIEVPGFPL
* .. ::: *.. :. : * * ** :*:. . : :. *
MMAR_2782|M.marinum_M INPGLSVTGHPSGTAKDVEETMHFAVLSGVRAWIEERPLAQAADGYEAME
MUL_2965|M.ulcerans_Agy99 INPGLSVTGHPSGTAKDVEDTMHFAVLSGVRAWIEERPLAQAADGYEAME
MAV_2812|M.avium_104 ITPGLSIVGHPSGTAKDVEDTMQFAVLSGVRAWIEELPLSRAADGYAALE
Mflv_0236|M.gilvum_PYR-GCK INDGLSVTGHPSGTSRDVEETLHFAMLSGVRARVEEVPLAQPAEAYEAMD
Mvan_0669|M.vanbaalenii_PYR-1 INAGRSVTGHPSGTSRDVEETMHFALLSGVRAKVEEVPLARAAEAYESMD
MSMEG_3464|M.smegmatis_MC2_155 IQPAVSISGHPSGTAREVEETMHFAVVAGVRARVEERPLDDAAEAYGAMA
Mb3071|M.bovis_AF2122/97 ALMRRSLAGSNIGGIAETQEMLNFCAEHGVTPEIELIEPDYINDAYERVL
Rv3045|M.tuberculosis_H37Rv ALMRRSLAGSNIGGIAETQEMLNFCAEHGVTPEIELIEPDYINDAYERVL
MLBr_01730|M.leprae_Br4923 ALMRRSLAGSNIGGITETQEMMNFSAEHSVTPEIELIEPDYINEAYERVL
TH_4581|M.thermoresistible__bu VGGRRRLAGSLIGGIAETQEMLDFCAEHGVLPEIEVIEPEYINTAYERVL
MAB_3400|M.abscessus_ATCC_1997 LTNRRSISGSLIGGIPETQEMLDFCAKHGIGSEIEVIDASYINEAYERVV
: * * :.:: :.*. .: . :* .* :
MMAR_2782|M.marinum_M QGRTRYRTVLAM-----
MUL_2965|M.ulcerans_Agy99 QGRTRYRTVLAM-----
MAV_2812|M.avium_104 GGRAHYRTVLTM-----
Mflv_0236|M.gilvum_PYR-GCK SSRARYRMVLTV-----
Mvan_0669|M.vanbaalenii_PYR-1 SGRARYRMVLTM-----
MSMEG_3464|M.smegmatis_MC2_155 DGRARYRFVLTT-----
Mb3071|M.bovis_AF2122/97 ASDVRYRFVIDISAL--
Rv3045|M.tuberculosis_H37Rv ASDVRYRFVIDISAL--
MLBr_01730|M.leprae_Br4923 ASDVRYRFVINISSL--
TH_4581|M.thermoresistible__bu ASDVRYRFVIDTASLRG
MAB_3400|M.abscessus_ATCC_1997 ASDVRYRFVIDTQTL--
. .:** *: