For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAELGTRERILQATAELYRRQGMSATGLKQISGAAKAPFGSIYHHFPGGKEAISVEVIRSEGIRYGEFVG AQLADTDPATGIPQLFESAGRLLQSQDYSEACSIETIALEVASTNEVLRQESATVFENWLTGLARWFGQL DVDEATSRRLAVITLTALEGAFVLCRTLRSVEPIIAAGQGVQAAVTATLAEKR
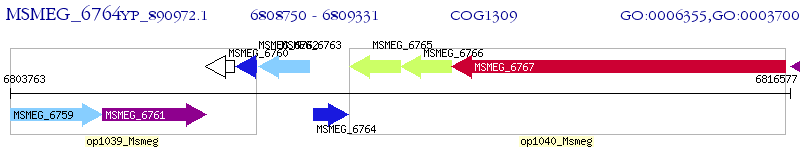
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6764 | - | - | 100% (193) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_3360 | - | 5e-17 | 27.62% (181) | transcriptional regulator, putative |
| M. smegmatis MC2 155 | MSMEG_3520 | - | 4e-15 | 25.52% (192) | TetR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0202 | - | 7e-16 | 27.32% (194) | putative transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3237 | - | 3e-77 | 76.84% (190) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0196 | - | 8e-16 | 27.32% (194) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2447c | - | 2e-41 | 49.73% (187) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0470 | - | 9e-23 | 31.15% (183) | transcriptional regulatory protein |
| M. avium 104 | MAV_0149 | - | 1e-33 | 42.62% (183) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3160 | - | 2e-15 | 34.91% (169) | PUTATIVE putative regulatory protein, TetR |
| M. ulcerans Agy99 | MUL_1119 | - | 2e-23 | 31.69% (183) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5860 | - | 1e-81 | 78.76% (193) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0202|M.bovis_AF2122/97 MQ-----------GPRERMVVSAALLIRERGAHATAISDVLQHSGAPRGS
Rv0196|M.tuberculosis_H37Rv MQ-----------GPRERMVVSAALLIRERGAHATAISDVLQHSGAPRGS
MMAR_0470|M.marinum_M MTPPSTSTPK-RGGTREKMLISAAQVMRERGAAGVTIDSVLARSGAPRGS
MUL_1119|M.ulcerans_Agy99 MTPPSTSTPK-RGGTREKMLISAAQVMRERGAAGVTIDSVLARSGAPRGS
TH_3160|M.thermoresistible__bu MGVEVTRRRDPQSGPRAAMLRSAVALMRRQGVAATSFADVLADSGAPRGS
MSMEG_6764|M.smegmatis_MC2_155 ---------MAELGTRERILQATAELYRRQGMSATGLKQISGAAKAPFGS
Mvan_5860|M.vanbaalenii_PYR-1 ---------MADPSTRSRILDATAELYRRQGMPATGLKQISAAAQAPFGS
Mflv_3237|M.gilvum_PYR-GCK ---------MTEPTTRDRILDAAAELYRRQGMSATGLKQISAAARAPFGS
MAB_2447c|M.abscessus_ATCC_199 ----------MPANTRDRIVAATCELFRRQGMTGTGLKQIAQSAGAPFGS
MAV_0149|M.avium_104 ----------MATATRERFLTAATGLFRRQGYSGTGLKQIVAESRAPLGS
.* :: :: : *.:* .. : .: : ** **
Mb0202|M.bovis_AF2122/97 AYHYFPGGRTQLLCEAVDYAGEHVAAMINEA--EGGLE----LLDALIDK
Rv0196|M.tuberculosis_H37Rv AYHYFPGGRTQLLCEAVDYAGEHVAAMINEA--EGGLE----LLDALIDK
MMAR_0470|M.marinum_M VYYHFPDGRNQILIEALRYAGDSITAVIDKAADKGARA----LLREFVRL
MUL_1119|M.ulcerans_Agy99 VYYHFPDGRNQILIEALRYAGDSITAVIDKAADKGARA----LLREFVRL
TH_3160|M.thermoresistible__bu IYHHFPAGKAQLIEEATRTAAAQLTERVARVLDEAADL--PTALRELVGV
MSMEG_6764|M.smegmatis_MC2_155 IYHHFPGGKEAISVEVIRSEGIRYGEFVGAQLADTDP--A-TGIPQLFES
Mvan_5860|M.vanbaalenii_PYR-1 IYHHFPGGKEAISVEVIRREGIRYAELVGAQLADTDP--L-TGIPQLFEK
Mflv_3237|M.gilvum_PYR-GCK IYHHFPGGKEAISAEVIRSEGIRYAAFVGAQLADINP--A-NGIEAVFAS
MAB_2447c|M.abscessus_ATCC_199 IYHFFPGGKAQLADEAIRAAGAMYRDLVLAVFDQNGPDLA-ATIRTAFAA
MAV_0149|M.avium_104 LYHFFPGGKQDLAVQAIAHTAERYRELLDRVFAKSTD-IG-QATVTWFNW
*:.** *: : :. . : . .
Mb0202|M.bovis_AF2122/97 YRQQLLSTDFRAGCPIAAVSVEAGDEQDRERMAPVIARAAAVFDRWSDLT
Rv0196|M.tuberculosis_H37Rv YRQQLLSTDFRAGCPIAAVSVEAGDEQDRERMAPVIARAAAVFDRWSDLT
MMAR_0470|M.marinum_M WEHLLTDGGFAAGCPVVAAALGPADDE-----VELAAEAGTILDSWCAAL
MUL_1119|M.ulcerans_Agy99 WEHLLTDGGFAAGCPVVAAALGPADDE-----VELAAEAGTILDSWCAAL
TH_3160|M.thermoresistible__bu WRDGLEAGDYAVGCPIVAAALGTER--------AARDVAGVAFADLTGLI
MSMEG_6764|M.smegmatis_MC2_155 AGRLLQSQDYSEACSIETIALEVASTN-----EVLRQESATVFENWLTGL
Mvan_5860|M.vanbaalenii_PYR-1 AGRDLEAQDYSEACSIETIALEVASTN-----ERLRVEAAEVFQSWLTGL
Mflv_3237|M.gilvum_PYR-GCK AAELLESQDYSEACSIETIALEVASTN-----ERLRGEAAEVFETWLHQL
MAB_2447c|M.abscessus_ATCC_199 AADNLIATDYTDACPIATIALEVASTD-----EDLRHATAEVFTDWIEQG
MAV_0149|M.avium_104 AARALQETDYADGCPIGTVACEVASSN-----EALRQATQAVLASWQSRV
* .: .*.: : : : :
Mb0202|M.bovis_AF2122/97 AQRFIADGIPPDRAHELAVLATSTLEGAILLARVRRDLTPLDLVHRQLRN
Rv0196|M.tuberculosis_H37Rv AQRFIADGIPPDRAHELAVLATSTLEGAILLARVRRDLTPLDLVHRQLRN
MMAR_0470|M.marinum_M GRAFVADGFDESTAASLAVTSIAALEGAIMLCRSTRSAEPLRQVGEQLEF
MUL_1119|M.ulcerans_Agy99 GRAFVADGFDESTAASLAVTSIAALEGAIMLCRSTRSVEPLRQVGEQLEF
TH_3160|M.thermoresistible__bu SDKLVSDGMARRRANSVAVLILTAMEGALVLAQARRDPAALDAVVDELTL
MSMEG_6764|M.smegmatis_MC2_155 ARWFGQLDVDEATSRRLAVITLTALEGAFVLCRTLRSVEPIIAAGQGVQA
Mvan_5860|M.vanbaalenii_PYR-1 AAWFGRLALTEPECRRLAMITLTALEGAFVLCRTLRSIDPIIAAGHGVQA
Mflv_3237|M.gilvum_PYR-GCK AGWFGQLDIGDEQSRRLATVMLTALEGAFVLCRTLRSVEPMLIAGHGVQA
MAB_2447c|M.abscessus_ATCC_199 TERIADTGLPPGTRRRLILGFITGLEGAFVLSRALRDPEPLFAAGETVAA
MAV_0149|M.avium_104 AAELIGAGVPTAQARRLATFAVASLEGAILLARAHRSTRPLRETGRVVAD
: . : : :***::*.: *. .: . :
Mb0202|M.bovis_AF2122/97 LLLAE-LPERSR----
Rv0196|M.tuberculosis_H37Rv LLLAE-LPERSR----
MMAR_0470|M.marinum_M LVKARGFVAQSRLDSE
MUL_1119|M.ulcerans_Agy99 LVKARGFVAQSRLDSE
TH_3160|M.thermoresistible__bu LCSGS---AD------
MSMEG_6764|M.smegmatis_MC2_155 AVTATLAEKR------
Mvan_5860|M.vanbaalenii_PYR-1 AVRAAATTRRS-----
Mflv_3237|M.gilvum_PYR-GCK AVHAVLASAQ------
MAB_2447c|M.abscessus_ATCC_199 AATSALAALATHAE--
MAV_0149|M.avium_104 TLRAATDRG-------
.