For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDPTGRTRLLGVVGDPVAQVRAPELWGTVFRELGVDAVCVPFHVRPTDLEAFLDGARRWRNLDGLLITVP HKAAAMRFATELSDRARQVGAVNCLKVGESGGWVADMLDGVGFVTAFEGGIGPVGGRRALVVGCGGAGSA IALALADAGASAVDVSDVDGARANELADRVSATGVLSRPVAALGRGYDLIVNASPAGMRSTDPLPVDLDG VGGDAAVADVITEPAVTPLIKEATQRGCAVQVGSQMTDAQVGGMADFFGYTAAFADPLR*
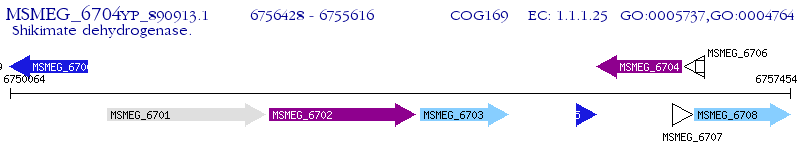
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6704 | - | - | 100% (270) | shikimate 5-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3028 | aroE | 3e-17 | 30.77% (260) | shikimate 5-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5457 | - | 1e-08 | 27.63% (228) | shikimate 5-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2582c | aroE | 1e-17 | 32.52% (246) | shikimate 5-dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3760 | aroE | 4e-16 | 30.74% (257) | shikimate 5-dehydrogenase |
| M. tuberculosis H37Rv | Rv2552c | aroE | 4e-17 | 32.78% (241) | shikimate 5-dehydrogenase |
| M. leprae Br4923 | MLBr_00515 | aroE | 2e-17 | 32.79% (244) | shikimate 5-dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2848c | aroE | 7e-18 | 31.42% (261) | shikimate 5-dehydrogenase |
| M. marinum M | MMAR_2173 | aroE | 1e-14 | 31.60% (250) | shikimate 5-dehydrogenase AroE |
| M. avium 104 | MAV_3428 | aroE | 2e-15 | 31.97% (244) | shikimate 5-dehydrogenase |
| M. thermoresistible (build 8) | TH_0111 | aroE | 1e-17 | 28.97% (252) | PROBABLE SHIKIMATE 5-DEHYDROGENASE AROE (5-DEHYDROSHIKIMATE |
| M. ulcerans Agy99 | MUL_1757 | aroE | 2e-14 | 33.14% (175) | shikimate 5-dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2643 | aroE | 1e-17 | 32.54% (252) | shikimate 5-dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2582c|M.bovis_AF2122/97 MSSTARDERR-------------------SQKAGVLGSPIAHSRSPQLHL
Rv2552c|M.tuberculosis_H37Rv MSEGPK-------------------------KAGVLGSPIAHSRSPQLHL
MLBr_00515|M.leprae_Br4923 MSLTACANKG-------------------PRKAGIIGSPIAHSRSPHLHL
MAV_3428|M.avium_104 MSSTAPPASGDRKRGGAGRSGSPPGQAAGPRRAAVLGKPIAHSKSPQLHL
MMAR_2173|M.marinum_M MSSGQEHSQPPR-----------------GRKAAVLGSPIAHSRSPQLHL
MUL_1757|M.ulcerans_Agy99 MSSGQEHNQPPR-----------------GRKAAVLGSPIAHSRSPQLHL
Mflv_3760|M.gilvum_PYR-GCK MSSTPAADRPRR--------------------AAVLGSPIAHSRSPQLHL
Mvan_2643|M.vanbaalenii_PYR-1 MMS----AGPRR--------------------AAVLGSPIAHSRSPQLHL
TH_0111|M.thermoresistible__bu ----------------------------------VLGSPIAHSRSPQLHL
MAB_2848c|M.abscessus_ATCC_199 MLAEIR-------------------------RAAVLGSPIGHSRSPDLHV
MSMEG_6704|M.smegmatis_MC2_155 MTDPTGRTRLLG----------------------VVGDPVAQVRAPELWG
::*.*:.: ::*.*
Mb2582c|M.bovis_AF2122/97 AAYRALGLHDWTYERIECGAAELPVVVGGFG--PEWVGVSVTMPGKFAAL
Rv2552c|M.tuberculosis_H37Rv AAYRALGLHDWTYERIECGAAELPVVVGGFG--PEWVGVSVTMPGKFAAL
MLBr_00515|M.leprae_Br4923 AAYRALRLHDWTYERIECDAEELPTVVSGFG--PQWVGVSVTMPGKFAAL
MAV_3428|M.avium_104 AAYRALGLHDWTYERIECDADQLPGVVGGFG--PEWVGVSVTMPGKFAAL
MMAR_2173|M.marinum_M AAYRALGLTDWTYQRIECDAAQLPAVVGGFG--PEWVGVSVTMPGKFAAL
MUL_1757|M.ulcerans_Agy99 AAYRALGLTDWTYQRIECDAAQLPAVVGGFG--PEWVGVSVTMPGKFAAL
Mflv_3760|M.gilvum_PYR-GCK AAYAALGLSSWRYDRIECGEDELPGFVAGLG--DEWVGLSVTMPGKFAAL
Mvan_2643|M.vanbaalenii_PYR-1 AAYRALGLTDWTYERIECTGEQLPGVVAGFG--PEWVGVSVTMPGKFAAL
TH_0111|M.thermoresistible__bu AAYRALGLTGWTYERIECDAEQLPVLVGGFG--PEWVGVSVTMPGKFAAL
MAB_2848c|M.abscessus_ATCC_199 AAYRALGLTGWTYERIECTAEQLPDVVGGAG--PEWVGFSVTMPGKFAAL
MSMEG_6704|M.smegmatis_MC2_155 TVFRELGVDAVCVP-FHVRPTDLEAFLDGARRWRNLDGLLITVPHKAAAM
:.: * : :. :* .: * : *. :*:* * **:
Mb2582c|M.bovis_AF2122/97 RFADERTARADLVGSANTLVR-TPHGWRADNTDIDGVAGALGAA-----A
Rv2552c|M.tuberculosis_H37Rv RFADERTARADLVGSANTLVR-TPHGWRADNTDIDGVAGALGAA-----A
MLBr_00515|M.leprae_Br4923 RFADEHTARASLVGSANTLLR-TQRGWRADNTDIDGVAGALAAIG--PLA
MAV_3428|M.avium_104 RFADERTERARRVGSANTLVR-TATGWRADNTDIDGVAGALGAA-----S
MMAR_2173|M.marinum_M QFADERTARAELVGSANTLVR-TPRGWRADNTDIDGVAGALGQA-----S
MUL_1757|M.ulcerans_Agy99 QFADERTARAELVGSANTLVR-TPRGWRADNTDIDGVAGALGQA-----S
Mflv_3760|M.gilvum_PYR-GCK RCADEATERARLVGSANTLVH-TATGWRADNTDIDGVAGALADA---GVS
Mvan_2643|M.vanbaalenii_PYR-1 RVADEATDRARMVGSANTLVR-SSAGWRADNTDIDGVVGALSGV---QVT
TH_0111|M.thermoresistible__bu TFADQRTARAELVGSANTLVR-RDGGWLADNTDIDGVKGALG-----AVG
MAB_2848c|M.abscessus_ATCC_199 RFASTSTERARSIGAANTLVR-SGDGWHADNTDVDGVSGALAGAGIDPAG
MSMEG_6704|M.smegmatis_MC2_155 RFATELSDRARQVGAVNCLKVGESGGWVADMLDGVGFVTAFEGGIGPVGG
* : ** :*:.* * ** ** * *. *:
Mb2582c|M.bovis_AF2122/97 GHALVLGSGGTAPAAVVGLAELGVTDITVVARNSDKAARLVDLGTRVGVA
Rv2552c|M.tuberculosis_H37Rv GHALVLGSGGTAPAAVVGLAELGVTDITVVARNSDKAARLVDLGTRVGVA
MLBr_00515|M.leprae_Br4923 GRALVCGSGGTAPAAVMGLAELGVTDITVLARNPDKASRLVDLGVQVGVA
MAV_3428|M.avium_104 GWALVCGSGGTAPAAVAGLAQLGVAGITVVARNPDKAARLVRLGDELGVP
MMAR_2173|M.marinum_M GPAIVCGSGGTAPAAVVGLAELGVTEITIAARNADKAARLVDLGARLGVA
MUL_1757|M.ulcerans_Agy99 GPAIVCGSGGTAPAAVVGLAELGVTEITIAARNADKAARLVDLGARLGVA
Mflv_3760|M.gilvum_PYR-GCK GSAVVVGSGGTAPAVVAGLVQLGAHHVTVVARNAEKAQGLVALGRRCGAT
Mvan_2643|M.vanbaalenii_PYR-1 DRAMVIGSGGTAPAVVVGLVTLGARFVTVAARHADKASALVELAVRCGAR
TH_0111|M.thermoresistible__bu EHVMVVGSGGTAPAAVVAVVESGARQVTVAARNREKAAPLVELAERLGAS
MAB_2848c|M.abscessus_ATCC_199 QSAVIVGAGGTARPAIVALAAMGVRALTVVARDAGRAQGVLELAEHLGLR
MSMEG_6704|M.smegmatis_MC2_155 RRALVVGCGGAGSAIALALADAGASAVDVSDVDGARANELADRVSATGVL
.:: *.**:. . .:. *. : : . :* : *
Mb2582c|M.bovis_AF2122/97 TRFCAFDSGGLADAVAAAEVLVSTIPAEVAAGYAGTLAAIPVLLDAIYDP
Rv2552c|M.tuberculosis_H37Rv TRFCAFDSGGLADAVAAAEVLVSTIPAEVAAGYAGTLAAIPVLLDAIYDP
MLBr_00515|M.leprae_Br4923 TRLCGLDSGGLADEVKAAEVLVSTVPADVAARYVDVFATVPVLLDAIYDP
MAV_3428|M.avium_104 TRFCALDGAGLADEVAAAEVLVSTLPAEVAERYAGTLARVGVLLDAVYDP
MMAR_2173|M.marinum_M SRFCGLDDPELGERAASAAALVSTIPAEVASRYAATFATVPVLLDAIYNP
MUL_1757|M.ulcerans_Agy99 SRFCGLDDPELGERAASAAALVSTIPAEVASRYAATFATVPVVLDAIYNS
Mflv_3760|M.gilvum_PYR-GCK TEWVDLTGDDVTGAVAAADVVVNTVPADAVAPYAPRLAGTPVLLDAIYDP
Mvan_2643|M.vanbaalenii_PYR-1 AGWVDLQHGRLAEEVAGVEVVVNTVPADAVAPHAAALAGTPVLLDAIYDP
TH_0111|M.thermoresistible__bu ARWQDLAG----VTAAGVDAVVSTIPADAAARYAAALSATPVLVDAIYDP
MAB_2848c|M.abscessus_ATCC_199 ASVLSFTDAELSSVCQAAGVLVSTVPAEAAAPYARSLAAAPAILDVIYHP
MSMEG_6704|M.smegmatis_MC2_155 SRPVAALGRGYDLIVNASPAGMRST--DPLPVDLDGVGGDAAVADVITEP
: . . : : : .. .: *.: ..
Mb2582c|M.bovis_AF2122/97 WPTPLAAAVGSAGGRVISGLQMLLHQAFAQVEQFTGLPAPREAMTCALAA
Rv2552c|M.tuberculosis_H37Rv WPTPLAAAVGSAGGRVISGLQMLLHQAFAQVEQFTGLPAPREAMTCALAA
MLBr_00515|M.leprae_Br4923 WPTPLVAAVSAAGGRVISGLQMLLHQAFAQVEQFTGMPAPREAMACALAG
MAV_3428|M.avium_104 WPTPLAAAVSAAGGRVISGMQMLLHQAFAQVEQFTGLPAPREAMTCAAAD
MMAR_2173|M.marinum_M WPTPLAAAVAAAGGRVISGLHMLLHQAFAQVEQFTGLPAPRAAMTCALEA
MUL_1757|M.ulcerans_Agy99 WPTPLAAAVAAAGGRVISGLHMLLHQAFAQVEQFTGLPAPRAAMTCALEA
Mflv_3760|M.gilvum_PYR-GCK WPTPLAAAVTAAGGRVISGLEMLLNQAFAQVEQFTGMPAPQKAMRGALGL
Mvan_2643|M.vanbaalenii_PYR-1 WPTPLAAAVQAAGGQVISGLEMLLNQAFTQVEQFTGLPAPKEAMRAALES
TH_0111|M.thermoresistible__bu WPTPLAAAVAEAGGRVISGLEMLLNQAFTQVELFTGRPAPKEAMRAALNQ
MAB_2848c|M.abscessus_ATCC_199 WPTTLAAAAQRRGAVVVGGLDMLLNQAFTQVELFTGLSAPRAAMAAALH-
MSMEG_6704|M.smegmatis_MC2_155 AVTPLIKEATQRGCAVQVGSQMTDAQVGGMADFFGYTAAFADPLR-----
*.* . * * * .* *. .: * .* .:
Mb2582c|M.bovis_AF2122/97 LD
Rv2552c|M.tuberculosis_H37Rv LD
MLBr_00515|M.leprae_Br4923 LH
MAV_3428|M.avium_104 AD
MMAR_2173|M.marinum_M LD
MUL_1757|M.ulcerans_Agy99 LD
Mflv_3760|M.gilvum_PYR-GCK A-
Mvan_2643|M.vanbaalenii_PYR-1 A-
TH_0111|M.thermoresistible__bu P-
MAB_2848c|M.abscessus_ATCC_199 --
MSMEG_6704|M.smegmatis_MC2_155 --