For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAEIRRAAVLGSPIGHSRSPDLHVAAYRALGLTGWTYERIECTAEQLPDVVGGAGPEWVGFSVTMPGKFA ALRFASTSTERARSIGAANTLVRSGDGWHADNTDVDGVSGALAGAGIDPAGQSAVIVGAGGTARPAIVAL AAMGVRALTVVARDAGRAQGVLELAEHLGLRASVLSFTDAELSSVCQAAGVLVSTVPAEAAAPYARSLAA APAILDVIYHPWPTTLAAAAQRRGAVVVGGLDMLLNQAFTQVELFTGLSAPRAAMAAALH*
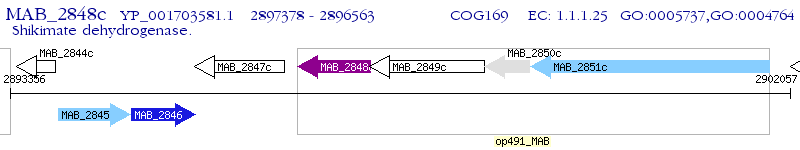
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2848c | aroE | - | 100% (271) | shikimate 5-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2582c | aroE | 4e-83 | 60.00% (265) | shikimate 5-dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3760 | aroE | 5e-83 | 57.74% (265) | shikimate 5-dehydrogenase |
| M. tuberculosis H37Rv | Rv2552c | aroE | 3e-83 | 60.00% (265) | shikimate 5-dehydrogenase |
| M. leprae Br4923 | MLBr_00515 | aroE | 5e-80 | 55.85% (265) | shikimate 5-dehydrogenase |
| M. marinum M | MMAR_2173 | aroE | 5e-84 | 58.87% (265) | shikimate 5-dehydrogenase AroE |
| M. avium 104 | MAV_3428 | aroE | 4e-82 | 58.71% (264) | shikimate 5-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3028 | aroE | 7e-87 | 60.45% (268) | shikimate 5-dehydrogenase |
| M. thermoresistible (build 8) | TH_0111 | aroE | 3e-82 | 60.98% (264) | PROBABLE SHIKIMATE 5-DEHYDROGENASE AROE (5-DEHYDROSHIKIMATE |
| M. ulcerans Agy99 | MUL_1757 | aroE | 2e-83 | 58.49% (265) | shikimate 5-dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2643 | aroE | 3e-88 | 61.13% (265) | shikimate 5-dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3028|M.smegmatis_MC2_155 MPGRRPAAPGE------------------PRKAAVLGSPIAHSRSPQLHL
TH_0111|M.thermoresistible__bu ----------------------------------VLGSPIAHSRSPQLHL
Mb2582c|M.bovis_AF2122/97 MSSTARDERR-------------------SQKAGVLGSPIAHSRSPQLHL
Rv2552c|M.tuberculosis_H37Rv MSEGPK-------------------------KAGVLGSPIAHSRSPQLHL
MLBr_00515|M.leprae_Br4923 MSLTACANKG-------------------PRKAGIIGSPIAHSRSPHLHL
MAV_3428|M.avium_104 MSSTAPPASGDRKRGGAGRSGSPPGQAAGPRRAAVLGKPIAHSKSPQLHL
MMAR_2173|M.marinum_M MSSGQEHSQPPR-----------------GRKAAVLGSPIAHSRSPQLHL
MUL_1757|M.ulcerans_Agy99 MSSGQEHNQPPR-----------------GRKAAVLGSPIAHSRSPQLHL
Mflv_3760|M.gilvum_PYR-GCK MSSTPAADRP--------------------RRAAVLGSPIAHSRSPQLHL
Mvan_2643|M.vanbaalenii_PYR-1 MMS----AGP--------------------RRAAVLGSPIAHSRSPQLHL
MAB_2848c|M.abscessus_ATCC_199 MLAEIR-------------------------RAAVLGSPIGHSRSPDLHV
::*.**.**:**.**:
MSMEG_3028|M.smegmatis_MC2_155 AAYRALGLTDWTYERIECTAEQLPGLVEAAGPEWVGFSVTMPGKFAALQF
TH_0111|M.thermoresistible__bu AAYRALGLTGWTYERIECDAEQLPVLVGGFGPEWVGVSVTMPGKFAALTF
Mb2582c|M.bovis_AF2122/97 AAYRALGLHDWTYERIECGAAELPVVVGGFGPEWVGVSVTMPGKFAALRF
Rv2552c|M.tuberculosis_H37Rv AAYRALGLHDWTYERIECGAAELPVVVGGFGPEWVGVSVTMPGKFAALRF
MLBr_00515|M.leprae_Br4923 AAYRALRLHDWTYERIECDAEELPTVVSGFGPQWVGVSVTMPGKFAALRF
MAV_3428|M.avium_104 AAYRALGLHDWTYERIECDADQLPGVVGGFGPEWVGVSVTMPGKFAALRF
MMAR_2173|M.marinum_M AAYRALGLTDWTYQRIECDAAQLPAVVGGFGPEWVGVSVTMPGKFAALQF
MUL_1757|M.ulcerans_Agy99 AAYRALGLTDWTYQRIECDAAQLPAVVGGFGPEWVGVSVTMPGKFAALQF
Mflv_3760|M.gilvum_PYR-GCK AAYAALGLSSWRYDRIECGEDELPGFVAGLGDEWVGLSVTMPGKFAALRC
Mvan_2643|M.vanbaalenii_PYR-1 AAYRALGLTDWTYERIECTGEQLPGVVAGFGPEWVGVSVTMPGKFAALRV
MAB_2848c|M.abscessus_ATCC_199 AAYRALGLTGWTYERIECTAEQLPDVVGGAGPEWVGFSVTMPGKFAALRF
*** ** * .* *:**** :** .* . * :***.***********
MSMEG_3028|M.smegmatis_MC2_155 ADQRTDRAELVGSANTLVRTEAGGWRADNTDVDGVAGALGTA-----GEA
TH_0111|M.thermoresistible__bu ADQRTARAELVGSANTLVRRD-GGWLADNTDIDGVKGALGAV-----GEH
Mb2582c|M.bovis_AF2122/97 ADERTARADLVGSANTLVRTP-HGWRADNTDIDGVAGALGAA-----AGH
Rv2552c|M.tuberculosis_H37Rv ADERTARADLVGSANTLVRTP-HGWRADNTDIDGVAGALGAA-----AGH
MLBr_00515|M.leprae_Br4923 ADEHTARASLVGSANTLLRTQ-RGWRADNTDIDGVAGALAAIGP--LAGR
MAV_3428|M.avium_104 ADERTERARRVGSANTLVRTA-TGWRADNTDIDGVAGALGAA-----SGW
MMAR_2173|M.marinum_M ADERTARAELVGSANTLVRTP-RGWRADNTDIDGVAGALGQA-----SGP
MUL_1757|M.ulcerans_Agy99 ADERTARAELVGSANTLVRTP-RGWRADNTDIDGVAGALGQA-----SGP
Mflv_3760|M.gilvum_PYR-GCK ADEATERARLVGSANTLVHTA-TGWRADNTDIDGVAGALADAG---VSGS
Mvan_2643|M.vanbaalenii_PYR-1 ADEATDRARMVGSANTLVRSS-AGWRADNTDIDGVVGALSGVQ---VTDR
MAB_2848c|M.abscessus_ATCC_199 ASTSTERARSIGAANTLVRSG-DGWHADNTDVDGVSGALAGAGIDPAGQS
*. * ** :*:****:: ** *****:*** ***.
MSMEG_3028|M.smegmatis_MC2_155 ALVIGSGGTAPAAVVGLAEMGVQHITIVARNADKAAALVDLAGRCGAQGD
TH_0111|M.thermoresistible__bu VMVVGSGGTAPAAVVAVVESGARQVTVAARNREKAAPLVELAERLGASAR
Mb2582c|M.bovis_AF2122/97 ALVLGSGGTAPAAVVGLAELGVTDITVVARNSDKAARLVDLGTRVGVATR
Rv2552c|M.tuberculosis_H37Rv ALVLGSGGTAPAAVVGLAELGVTDITVVARNSDKAARLVDLGTRVGVATR
MLBr_00515|M.leprae_Br4923 ALVCGSGGTAPAAVMGLAELGVTDITVLARNPDKASRLVDLGVQVGVATR
MAV_3428|M.avium_104 ALVCGSGGTAPAAVAGLAQLGVAGITVVARNPDKAARLVRLGDELGVPTR
MMAR_2173|M.marinum_M AIVCGSGGTAPAAVVGLAELGVTEITIAARNADKAARLVDLGARLGVASR
MUL_1757|M.ulcerans_Agy99 AIVCGSGGTAPAAVVGLAELGVTEITIAARNADKAARLVDLGARLGVASR
Mflv_3760|M.gilvum_PYR-GCK AVVVGSGGTAPAVVAGLVQLGAHHVTVVARNAEKAQGLVALGRRCGATTE
Mvan_2643|M.vanbaalenii_PYR-1 AMVIGSGGTAPAVVVGLVTLGARFVTVAARHADKASALVELAVRCGARAG
MAB_2848c|M.abscessus_ATCC_199 AVIVGAGGTARPAIVALAAMGVRALTVVARDAGRAQGVLELAEHLGLRAS
.:: *:**** ..: .:. *. :*: **. :* :: *. . *
MSMEG_3028|M.smegmatis_MC2_155 WCDIAGPALTDAVAQASAAVSTIPADAAAGYAPTLAAVPRLLDAIYNPWP
TH_0111|M.thermoresistible__bu WQDLAG----VTAAGVDAVVSTIPADAAARYAAALSATPVLVDAIYDPWP
Mb2582c|M.bovis_AF2122/97 FCAFDSGGLADAVAAAEVLVSTIPAEVAAGYAGTLAAIPVLLDAIYDPWP
Rv2552c|M.tuberculosis_H37Rv FCAFDSGGLADAVAAAEVLVSTIPAEVAAGYAGTLAAIPVLLDAIYDPWP
MLBr_00515|M.leprae_Br4923 LCGLDSGGLADEVKAAEVLVSTVPADVAARYVDVFATVPVLLDAIYDPWP
MAV_3428|M.avium_104 FCALDGAGLADEVAAAEVLVSTLPAEVAERYAGTLARVGVLLDAVYDPWP
MMAR_2173|M.marinum_M FCGLDDPELGERAASAAALVSTIPAEVASRYAATFATVPVLLDAIYNPWP
MUL_1757|M.ulcerans_Agy99 FCGLDDPELGERAASAAALVSTIPAEVASRYAATFATVPVVLDAIYNSWP
Mflv_3760|M.gilvum_PYR-GCK WVDLTGDDVTGAVAAADVVVNTVPADAVAPYAPRLAGTPVLLDAIYDPWP
Mvan_2643|M.vanbaalenii_PYR-1 WVDLQHGRLAEEVAGVEVVVNTVPADAVAPHAAALAGTPVLLDAIYDPWP
MAB_2848c|M.abscessus_ATCC_199 VLSFTDAELSSVCQAAGVLVSTVPAEAAAPYARSLAAAPAILDVIYHPWP
: . . *.*:**:.. :. :: ::*.:*..**
MSMEG_3028|M.smegmatis_MC2_155 TPLATAVQAAGGEVISGLQMLLNQAYSQVELFTGRPAPKEAMRAALEEP-
TH_0111|M.thermoresistible__bu TPLAAAVAEAGGRVISGLEMLLNQAFTQVELFTGRPAPKEAMRAALNQP-
Mb2582c|M.bovis_AF2122/97 TPLAAAVGSAGGRVISGLQMLLHQAFAQVEQFTGLPAPREAMTCALAALD
Rv2552c|M.tuberculosis_H37Rv TPLAAAVGSAGGRVISGLQMLLHQAFAQVEQFTGLPAPREAMTCALAALD
MLBr_00515|M.leprae_Br4923 TPLVAAVSAAGGRVISGLQMLLHQAFAQVEQFTGMPAPREAMACALAGLH
MAV_3428|M.avium_104 TPLAAAVSAAGGRVISGMQMLLHQAFAQVEQFTGLPAPREAMTCAAADAD
MMAR_2173|M.marinum_M TPLAAAVAAAGGRVISGLHMLLHQAFAQVEQFTGLPAPRAAMTCALEALD
MUL_1757|M.ulcerans_Agy99 TPLAAAVAAAGGRVISGLHMLLHQAFAQVEQFTGLPAPRAAMTCALEALD
Mflv_3760|M.gilvum_PYR-GCK TPLAAAVTAAGGRVISGLEMLLNQAFAQVEQFTGMPAPQKAMRGALGLA-
Mvan_2643|M.vanbaalenii_PYR-1 TPLAAAVQAAGGQVISGLEMLLNQAFTQVEQFTGLPAPKEAMRAALESA-
MAB_2848c|M.abscessus_ATCC_199 TTLAAAAQRRGAVVVGGLDMLLNQAFTQVELFTGLSAPRAAMAAALH---
*.*.:*. *. *:.*:.***:**::*** *** .**: ** *