For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGADNRRPPINKDTKVCISLAARPSNHGTRFHNYLYDKLGLDFLYKAFSTTDIAAAIGGVRALGIRGCSV SMPFKQDVMALVDEIEPSARAIDAVNTIVNDLSVPGGYLVASNTDYIAVQALIEQHGLAEDDPVLIRGSG GMANAVAAAFRDNGFRRGTIVARNRDTGAALASRLGYEYTPEVAERTAAILVNVTPIGMAGGPEAQLSAF EPGTVEAAAVVFDVVAMPAETPLIVAARSAGKPVITGAEVIALQAAEQFERYTGVRPTAEQIAEASAFSR A*
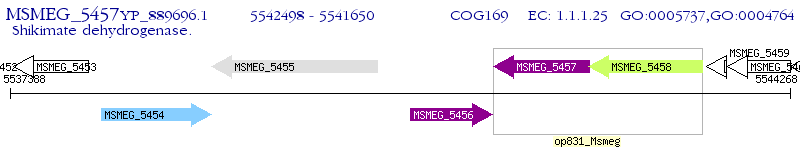
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5457 | - | - | 100% (282) | shikimate 5-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3028 | aroE | 2e-16 | 32.58% (264) | shikimate 5-dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6704 | - | 1e-08 | 27.63% (228) | shikimate 5-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2582c | aroE | 2e-16 | 30.77% (260) | shikimate 5-dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1916 | - | 1e-109 | 73.19% (276) | shikimate 5-dehydrogenase |
| M. tuberculosis H37Rv | Rv2552c | aroE | 2e-16 | 30.77% (260) | shikimate 5-dehydrogenase |
| M. leprae Br4923 | MLBr_00515 | aroE | 4e-13 | 27.03% (259) | shikimate 5-dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2848c | aroE | 8e-12 | 29.17% (264) | shikimate 5-dehydrogenase |
| M. marinum M | MMAR_2173 | aroE | 6e-19 | 30.47% (279) | shikimate 5-dehydrogenase AroE |
| M. avium 104 | MAV_3428 | aroE | 9e-16 | 30.94% (265) | shikimate 5-dehydrogenase |
| M. thermoresistible (build 8) | TH_0111 | aroE | 5e-17 | 32.57% (218) | PROBABLE SHIKIMATE 5-DEHYDROGENASE AROE (5-DEHYDROSHIKIMATE |
| M. ulcerans Agy99 | MUL_1757 | aroE | 1e-18 | 30.47% (279) | shikimate 5-dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4817 | - | 1e-104 | 70.55% (275) | shikimate 5-dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2582c|M.bovis_AF2122/97 ----------------MSSTARDERR-------------------SQKAG
Rv2552c|M.tuberculosis_H37Rv ----------------MSEGPK-------------------------KAG
MLBr_00515|M.leprae_Br4923 ----------------MSLTACANKG-------------------PRKAG
MAV_3428|M.avium_104 ----------------MSSTAPPASGDRKRGGAGRSGSPPGQAAGPRRAA
MMAR_2173|M.marinum_M ----------------MSSGQEHSQPPR-----------------GRKAA
MUL_1757|M.ulcerans_Agy99 ----------------MSSGQEHNQPPR-----------------GRKAA
TH_0111|M.thermoresistible__bu --------------------------------------------------
MAB_2848c|M.abscessus_ATCC_199 ----------------MLAEIR-------------------------RAA
Mflv_1916|M.gilvum_PYR-GCK MPRRRAADQTERRAADQTEQRARRPPLN----------------KDTRVC
Mvan_4817|M.vanbaalenii_PYR-1 ---------------------MTRPPLT----------------KDTRLC
MSMEG_5457|M.smegmatis_MC2_155 ----------------MSGADNRRPPIN----------------KDTKVC
Mb2582c|M.bovis_AF2122/97 VLGSPIAHSRSPQLHLAAYRALGLHDWTYERIECGAAELPVVVGGFGPEW
Rv2552c|M.tuberculosis_H37Rv VLGSPIAHSRSPQLHLAAYRALGLHDWTYERIECGAAELPVVVGGFGPEW
MLBr_00515|M.leprae_Br4923 IIGSPIAHSRSPHLHLAAYRALRLHDWTYERIECDAEELPTVVSGFGPQW
MAV_3428|M.avium_104 VLGKPIAHSKSPQLHLAAYRALGLHDWTYERIECDADQLPGVVGGFGPEW
MMAR_2173|M.marinum_M VLGSPIAHSRSPQLHLAAYRALGLTDWTYQRIECDAAQLPAVVGGFGPEW
MUL_1757|M.ulcerans_Agy99 VLGSPIAHSRSPQLHLAAYRALGLTDWTYQRIECDAAQLPAVVGGFGPEW
TH_0111|M.thermoresistible__bu VLGSPIAHSRSPQLHLAAYRALGLTGWTYERIECDAEQLPVLVGGFGPEW
MAB_2848c|M.abscessus_ATCC_199 VLGSPIGHSRSPDLHVAAYRALGLTGWTYERIECTAEQLPDVVGGAGPEW
Mflv_1916|M.gilvum_PYR-GCK ISLSGRPSNIGTRFHNHLYDVYGLD---FLYKAFTTTDIVAAIGGVRALG
Mvan_4817|M.vanbaalenii_PYR-1 ISLAGRPSNIGTRFHNHLYEVLGLD---FIYKAFTTTDIVAAIGGVRALG
MSMEG_5457|M.smegmatis_MC2_155 ISLAARPSNHGTRFHNYLYDKLGLD---FLYKAFSTTDIAAAIGGVRALG
: . .. :* * * : : :: :.* .
Mb2582c|M.bovis_AF2122/97 VG-VSVTMPGKFAALRFADERTARADLVGSANTLVRTP----HGWRADNT
Rv2552c|M.tuberculosis_H37Rv VG-VSVTMPGKFAALRFADERTARADLVGSANTLVRTP----HGWRADNT
MLBr_00515|M.leprae_Br4923 VG-VSVTMPGKFAALRFADEHTARASLVGSANTLLRTQ----RGWRADNT
MAV_3428|M.avium_104 VG-VSVTMPGKFAALRFADERTERARRVGSANTLVRTA----TGWRADNT
MMAR_2173|M.marinum_M VG-VSVTMPGKFAALQFADERTARAELVGSANTLVRTP----RGWRADNT
MUL_1757|M.ulcerans_Agy99 VG-VSVTMPGKFAALQFADERTARAELVGSANTLVRTP----RGWRADNT
TH_0111|M.thermoresistible__bu VG-VSVTMPGKFAALTFADQRTARAELVGSANTLVRRD----GGWLADNT
MAB_2848c|M.abscessus_ATCC_199 VG-FSVTMPGKFAALRFASTSTERARSIGAANTLVRSG----DGWHADNT
Mflv_1916|M.gilvum_PYR-GCK IRGCSVSMPFKEDVLALVDEIEPSAQAIRSANTIVNDVSVPGGRLTASNT
Mvan_4817|M.vanbaalenii_PYR-1 IRGCSVSMPFKEDVLHLVDHVEASAQAIESVNTIVNDD----GRLTASNT
MSMEG_5457|M.smegmatis_MC2_155 IRGCSVSMPFKQDVMALVDEIEPSARAIDAVNTIVNDLSVPGGYLVASNT
: **:** * .: :.. * : :.**::. *.**
Mb2582c|M.bovis_AF2122/97 DIDGVAGALGAA-----AGHALVLGSGGTAPAAVVGLAELGVTDITVVAR
Rv2552c|M.tuberculosis_H37Rv DIDGVAGALGAA-----AGHALVLGSGGTAPAAVVGLAELGVTDITVVAR
MLBr_00515|M.leprae_Br4923 DIDGVAGALAAIGPL--AGRALVCGSGGTAPAAVMGLAELGVTDITVLAR
MAV_3428|M.avium_104 DIDGVAGALGAA-----SGWALVCGSGGTAPAAVAGLAQLGVAGITVVAR
MMAR_2173|M.marinum_M DIDGVAGALGQA-----SGPAIVCGSGGTAPAAVVGLAELGVTEITIAAR
MUL_1757|M.ulcerans_Agy99 DIDGVAGALGQA-----SGPAIVCGSGGTAPAAVVGLAELGVTEITIAAR
TH_0111|M.thermoresistible__bu DIDGVKGALGAV-----GEHVMVVGSGGTAPAAVVAVVESGARQVTVAAR
MAB_2848c|M.abscessus_ATCC_199 DVDGVSGALAGAGIDPAGQSAVIVGAGGTARPAIVALAAMGVRALTVVAR
Mflv_1916|M.gilvum_PYR-GCK DYLAVGQLIDDNGLDP-HDAVLIRGSGGMAKAVGAAFVDAGFRTGFVVAR
Mvan_4817|M.vanbaalenii_PYR-1 DYLAVQQLIGEHGLDP-EDTVMIRGSGGMAKAVGAAFAGAGFRTGFVVAR
MSMEG_5457|M.smegmatis_MC2_155 DYIAVQALIEQHGLAE-DDPVLIRGSGGMANAVAAAFRDNGFRRGTIVAR
* .* : .:: *:** * .. .. * : **
Mb2582c|M.bovis_AF2122/97 NSDKAARLVDLGTRVGVATRFCAFDSGGLADAVAAAEVLVSTIPAEVAAG
Rv2552c|M.tuberculosis_H37Rv NSDKAARLVDLGTRVGVATRFCAFDSGGLADAVAAAEVLVSTIPAEVAAG
MLBr_00515|M.leprae_Br4923 NPDKASRLVDLGVQVGVATRLCGLDSGGLADEVKAAEVLVSTVPADVAAR
MAV_3428|M.avium_104 NPDKAARLVRLGDELGVPTRFCALDGAGLADEVAAAEVLVSTLPAEVAER
MMAR_2173|M.marinum_M NADKAARLVDLGARLGVASRFCGLDDPELGERAASAAALVSTIPAEVASR
MUL_1757|M.ulcerans_Agy99 NADKAARLVDLGARLGVASRFCGLDDPELGERAASAAALVSTIPAEVASR
TH_0111|M.thermoresistible__bu NREKAAPLVELAERLGASARWQDLAG----VTAAGVDAVVSTIPADAAAR
MAB_2848c|M.abscessus_ATCC_199 DAGRAQGVLELAEHLGLRASVLSFTDAELSSVCQAAGVLVSTVPAEAAAP
Mflv_1916|M.gilvum_PYR-GCK NAATGA---ALADRLGYEYRPEVGSVEASVLVNVTPVGMAGGLDADEIAF
Mvan_4817|M.vanbaalenii_PYR-1 NPESGH---ALADRLGYTYVADVGTVTAPIIVNVTPIGMAGAPEQRELAF
MSMEG_5457|M.smegmatis_MC2_155 NRDTGA---ALASRLGYEYTPEVAERTAAILVNVTPIGMAGGPEAQLSAF
: . *. .:* :..
Mb2582c|M.bovis_AF2122/97 YAGTLAAIPVLLDAIYDPWPTPLAAAVGSAGGRVISGLQMLLHQAFAQVE
Rv2552c|M.tuberculosis_H37Rv YAGTLAAIPVLLDAIYDPWPTPLAAAVGSAGGRVISGLQMLLHQAFAQVE
MLBr_00515|M.leprae_Br4923 YVDVFATVPVLLDAIYDPWPTPLVAAVSAAGGRVISGLQMLLHQAFAQVE
MAV_3428|M.avium_104 YAGTLARVGVLLDAVYDPWPTPLAAAVSAAGGRVISGMQMLLHQAFAQVE
MMAR_2173|M.marinum_M YAATFATVPVLLDAIYNPWPTPLAAAVAAAGGRVISGLHMLLHQAFAQVE
MUL_1757|M.ulcerans_Agy99 YAATFATVPVVLDAIYNSWPTPLAAAVAAAGGRVISGLHMLLHQAFAQVE
TH_0111|M.thermoresistible__bu YAAALSATPVLVDAIYDPWPTPLAAAVAEAGGRVISGLEMLLNQAFTQVE
MAB_2848c|M.abscessus_ATCC_199 YARSLAAAPAILDVIYHPWPTTLAAAAQRRGAVVVGGLDMLLNQAFTQVE
Mflv_1916|M.gilvum_PYR-GCK PGSAIAAADTVFDVVAMPSETPLIRAARAADRRVITGAEVVALQAAEQFA
Mvan_4817|M.vanbaalenii_PYR-1 GAAAIAAADTVFDVVALPSETPLIKAARAAGAQVITGAQVIALQAAEQFE
MSMEG_5457|M.smegmatis_MC2_155 EPGTVEAAAVVFDVVAMPAETPLIVAARSAGKPVITGAEVIALQAAEQFE
. .:.*.: . *.* *. . *: * .:: ** *.
Mb2582c|M.bovis_AF2122/97 QFTGLPAPREAMTCALAALD-
Rv2552c|M.tuberculosis_H37Rv QFTGLPAPREAMTCALAALD-
MLBr_00515|M.leprae_Br4923 QFTGMPAPREAMACALAGLH-
MAV_3428|M.avium_104 QFTGLPAPREAMTCAAADAD-
MMAR_2173|M.marinum_M QFTGLPAPRAAMTCALEALD-
MUL_1757|M.ulcerans_Agy99 QFTGLPAPRAAMTCALEALD-
TH_0111|M.thermoresistible__bu LFTGRPAPKEAMRAALNQP--
MAB_2848c|M.abscessus_ATCC_199 LFTGLSAPRAAMAAALH----
Mflv_1916|M.gilvum_PYR-GCK RYTGVRPDPGHVAEASAVSRA
Mvan_4817|M.vanbaalenii_PYR-1 RYTGVRPTPEQIAEASALSRA
MSMEG_5457|M.smegmatis_MC2_155 RYTGVRPTAEQIAEASAFSRA
:** . : *