For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HATRRQALVGTGITLAATAVAGCTTYGDGPAKSDPSDSAGPAPGGGEVIAEVADVPVGSGVIVGDTVVTQ PTAGVFKGFSSKCTHKGCTVNEIADGTIHCPCHGSEFNLDGTVAKGPATRPLDAKSVEVEGDSIVLQ*
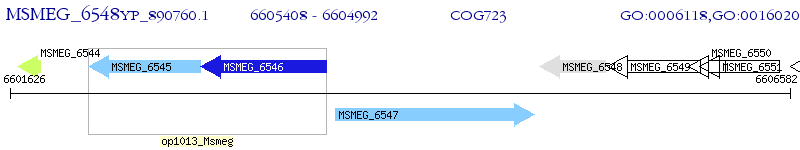
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6548 | - | - | 100% (138) | Rieske iron-sulfur protein |
| M. smegmatis MC2 155 | MSMEG_0637 | - | 5e-05 | 37.50% (72) | iron-sulfur binding oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4709 | - | 3e-30 | 67.47% (83) | Rieske (2Fe-2S) domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4653 | - | 4e-07 | 32.65% (147) | hypothetical protein MAB_4653 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4414 | - | 4e-08 | 28.76% (153) | Rieske (2Fe-2S) domain-containing protein |
| M. thermoresistible (build 8) | TH_4003 | - | 8e-07 | 31.40% (86) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3864 | - | 2e-40 | 56.74% (141) | Rieske (2Fe-2S) domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6548|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3864|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4709|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4653|M.abscessus_ATCC_1997 MNPRGLRRYVDDLLRGRRPKSFRPDDFEAAQIRTAIELRASRPGD---DA
MAV_4414|M.avium_104 MNARGLRRYVDDLLRGRRPRPFLPDDFEAAQLRTAIELRAARPDSEVADA
TH_4003|M.thermoresistible__bu --------------------------------------------------
MSMEG_6548|M.smegmatis_MC2_155 ----------------------------MHATRRQALVGTGITLAATAVA
Mvan_3864|M.vanbaalenii_PYR-1 ----------------------------MSLSRRHALLGTGITLAASAVA
Mflv_4709|M.gilvum_PYR-GCK ----------------------------MWRWRSPGRQCS----SARASA
MAB_4653|M.abscessus_ATCC_1997 PSQEFLAELGRRLAEQISDTPVSSAPRRWQAPRRTVLVGTSTAAAAAAVA
MAV_4414|M.avium_104 PRPEFLADLHRRLAEQMSGAPTRPRPGR-GSTRRQVIVGTSAAATAAVAA
TH_4003|M.thermoresistible__bu --------------------------------------------------
MSMEG_6548|M.smegmatis_MC2_155 GCTTYGD-----GPAKSDPSDSAGP--APGGGEVIAEVADVPVGSGVIVG
Mvan_3864|M.vanbaalenii_PYR-1 GCATYGRSV---TPAPAAP-ESPGPEETTETTAALAATADVPVGSGIVVG
Mflv_4709|M.gilvum_PYR-GCK SPRSRWRHA---REVRTRRDRRRGHLRRRARSWRRRPTSDVPVGSGTIVG
MAB_4653|M.abscessus_ATCC_1997 VTADHLMTP---TPQTDTHQAPGEIVPNTGSWQRVAVSNAVPDGGVHPFD
MAV_4414|M.avium_104 VSIDRTVIAGQSGPDRAPQIASGTLAPNQGSWQRVAASSEVPDGVLRAFD
TH_4003|M.thermoresistible__bu ---------------------MSGEHTDTLRWVRAACLDDVPGDEPLGLV
** . .
MSMEG_6548|M.smegmatis_MC2_155 DTVVT---QPTAGVFKGFSSKCTHKGCTVN--EIADGTIHCPCHGSEFNL
Mvan_3864|M.vanbaalenii_PYR-1 DVVLT---QPNTGEFKAFSAVCTHAGCTVS--EVIGASITCPCHGSSFHL
Mflv_4709|M.gilvum_PYR-GCK EVVVT---QPVAGEFKAFSAVCTHTGCLVN--KVADGTIDCPCHGSRFSL
MAB_4653|M.abscessus_ATCC_1997 LGFVNGFVRRVGGRVEAVSGVCTHQGCKLWF-DGTNNRLQCPCHTTSFTT
MAV_4414|M.avium_104 LGAVSGFVRRVDGKVQAISGVCTHQGCKLWF-DAPDDTLRCPCHTTSFSP
TH_4003|M.thermoresistible__bu VDGIEIALFNDDGRIFALADRCTHGDARLSDGYIEDGCIECPLHQALFDL
: * . ..: *** .. : . : ** * : *
MSMEG_6548|M.smegmatis_MC2_155 D-GTVAKG---PATRPLDAKSVEVEGDSIVLQ------
Mvan_3864|M.vanbaalenii_PYR-1 D-GTVATG---PAKRALDTTPITVQGDSITRA------
Mflv_4709|M.gilvum_PYR-GCK D-GAVVNG---PAEKPLQPVAVRVQGDSIVAD------
MAB_4653|M.abscessus_ATCC_1997 D-GRVITHQLPIAPKPLPTLQVRETEGHIEVFAPDRPV
MAV_4414|M.avium_104 S-GKVLTHQLPIEPKPLPALMVREADGIIEVFAPVRRT
TH_4003|M.thermoresistible__bu ATGEVRRG---PAFEPVRSFEIRIDGDDIIVGL-----
* * ..: . : . *