For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNPRGLRRYVDDLLRGRRPKSFRPDDFEAAQIRTAIELRASRPGDDAPSQEFLAELGRRLAEQISDTPVS SAPRRWQAPRRTVLVGTSTAAAAAAVAVTADHLMTPTPQTDTHQAPGEIVPNTGSWQRVAVSNAVPDGGV HPFDLGFVNGFVRRVGGRVEAVSGVCTHQGCKLWFDGTNNRLQCPCHTTSFTTDGRVITHQLPIAPKPLP TLQVRETEGHIEVFAPDRPV
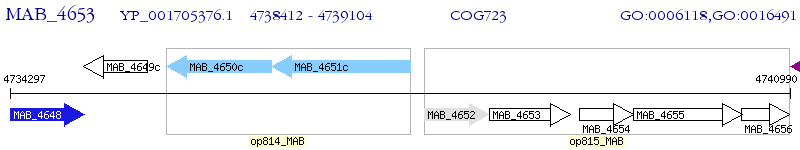
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4653 | - | - | 100% (230) | hypothetical protein MAB_4653 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4709 | - | 4e-08 | 31.51% (146) | Rieske (2Fe-2S) domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4414 | - | 1e-78 | 63.25% (234) | Rieske (2Fe-2S) domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_6548 | - | 1e-06 | 32.65% (147) | Rieske iron-sulfur protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3864 | - | 6e-09 | 35.20% (125) | Rieske (2Fe-2S) domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6548|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3864|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4709|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4653|M.abscessus_ATCC_1997 MNPRGLRRYVDDLLRGRRPKSFRPDDFEAAQIRTAIELRASRPGD---DA
MAV_4414|M.avium_104 MNARGLRRYVDDLLRGRRPRPFLPDDFEAAQLRTAIELRAARPDSEVADA
MSMEG_6548|M.smegmatis_MC2_155 ----------------------------MHATRRQALVGTGITLAATAVA
Mvan_3864|M.vanbaalenii_PYR-1 ----------------------------MSLSRRHALLGTGITLAASAVA
Mflv_4709|M.gilvum_PYR-GCK ----------------------------MWRWRSPGRQCS----SARASA
MAB_4653|M.abscessus_ATCC_1997 PSQEFLAELGRRLAEQISDTPVSSAPRRWQAPRRTVLVGTSTAAAAAAVA
MAV_4414|M.avium_104 PRPEFLADLHRRLAEQMSGAPTRPRPGR-GSTRRQVIVGTSAAATAAVAA
* : :* . *
MSMEG_6548|M.smegmatis_MC2_155 GCTTYGD-----GPAKSDPSDSAGP--APGGGEVIAEVADVPVGSGVIVG
Mvan_3864|M.vanbaalenii_PYR-1 GCATYGRSV---TPAPAAP-ESPGPEETTETTAALAATADVPVGSGIVVG
Mflv_4709|M.gilvum_PYR-GCK SPRSRWRHA---REVRTRRDRRRGHLRRRARSWRRRPTSDVPVGSGTIVG
MAB_4653|M.abscessus_ATCC_1997 VTADHLMTP---TPQTDTHQAPGEIVPNTGSWQRVAVSNAVPDGGVHPFD
MAV_4414|M.avium_104 VSIDRTVIAGQSGPDRAPQIASGTLAPNQGSWQRVAASSEVPDGVLRAFD
** * ..
MSMEG_6548|M.smegmatis_MC2_155 DTVVT---QPTAGVFKGFSSKCTHKGCTVN-EIADGTIHCPCHGSEFNLD
Mvan_3864|M.vanbaalenii_PYR-1 DVVLT---QPNTGEFKAFSAVCTHAGCTVS-EVIGASITCPCHGSSFHLD
Mflv_4709|M.gilvum_PYR-GCK EVVVT---QPVAGEFKAFSAVCTHTGCLVN-KVADGTIDCPCHGSRFSLD
MAB_4653|M.abscessus_ATCC_1997 LGFVNGFVRRVGGRVEAVSGVCTHQGCKLWFDGTNNRLQCPCHTTSFTTD
MAV_4414|M.avium_104 LGAVSGFVRRVDGKVQAISGVCTHQGCKLWFDAPDDTLRCPCHTTSFSPS
:. : * .:..*. *** ** : . . : **** : * .
MSMEG_6548|M.smegmatis_MC2_155 GTVAKG---PATRPLDAKSVEVEGDSIVLQ------
Mvan_3864|M.vanbaalenii_PYR-1 GTVATG---PAKRALDTTPITVQGDSITRA------
Mflv_4709|M.gilvum_PYR-GCK GAVVNG---PAEKPLQPVAVRVQGDSIVAD------
MAB_4653|M.abscessus_ATCC_1997 GRVITHQLPIAPKPLPTLQVRETEGHIEVFAPDRPV
MAV_4414|M.avium_104 GKVLTHQLPIEPKPLPALMVREADGIIEVFAPVRRT
* * . :.* . : . *