For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASIEIQELVKEFTDRHGAVTRVIDGIDLTISGETFVSVVGPSGSGKTTLLNIVSGIEPHTSGSVRLREG SRDARVGYVFQDPRLLPWRTVMANLGFVQHERTGCQERAEHYLDLVGLSHCADRFPAQLSGGQQQRIGIA RAFAVEPDVLLMDEPFSHLDAMTSRTLREHLERIWLESRRTVMFVTHDVTEAVQLSDRIVVLAPGGYIHE VIDVELPRPRRASDPAVAVLQAEVLARFEALEKRRTQLQDIVD
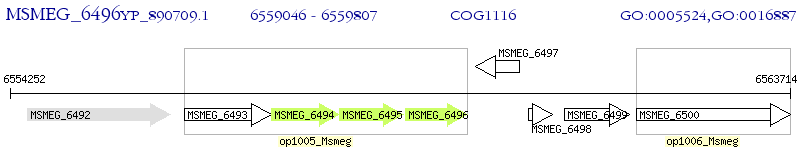
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6496 | - | - | 100% (253) | nitrate/sulfonate/bicarbonate ABC transporter ATPase subunit |
| M. smegmatis MC2 155 | MSMEG_2633 | - | 2e-45 | 44.30% (228) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_2459 | - | 4e-43 | 38.61% (259) | transporter ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3784c | proV | 2e-33 | 38.94% (208) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3158 | - | 1e-40 | 42.04% (226) | ABC transporter related |
| M. tuberculosis H37Rv | Rv3758c | proV | 2e-33 | 38.94% (208) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | MLBr_01424 | - | 1e-31 | 36.74% (215) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_2178 | - | 3e-37 | 40.69% (231) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_0034 | proV_1 | 8e-37 | 37.21% (258) | osmoprotectant (glycine |
| M. avium 104 | MAV_0140 | - | 8e-40 | 40.18% (224) | ABC nitrate/sulfonate/bicarbonate transporter, ATPase subunit |
| M. thermoresistible (build 8) | TH_4034 | - | 9e-37 | 37.96% (216) | PUTATIVE ABC transporter, ATP-binding protein SugC |
| M. ulcerans Agy99 | MUL_0033 | proV_1 | 1e-35 | 36.43% (258) | osmoprotectant (glycine |
| M. vanbaalenii PYR-1 | Mvan_0123 | - | 7e-38 | 43.32% (217) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3158|M.gilvum_PYR-GCK -----------------MSSPVVTVRGLHRAFGAQQ-----VLDDLDLDI
MAV_0140|M.avium_104 -----------------------------------------MLRDVDVEI
MSMEG_6496|M.smegmatis_MC2_155 -----------------MAS--IEIQELVKEFTDRHGAVTRVIDGIDLTI
MAB_2178|M.abscessus_ATCC_1997 ----MPLVDEAETAVQERDTGSIRISGVTHSYGSGAGKTT-ALGPVDLTV
Mvan_0123|M.vanbaalenii_PYR-1 --------------MPSTNDGAIRIDSVSHHYGSGGAEVT-ALGPVDLTV
MLBr_01424|M.leprae_Br4923 -------------------MASVTFEQATRQFPDTD---RPALDRLDLDV
TH_4034|M.thermoresistible__bu -------------------MAEIVLDKVTKSYPDGAGGVRHAVKDLSLTI
Mb3784c|M.bovis_AF2122/97 ---------------------MICFDDVSKVYAHGAT----AVDRLTLEV
Rv3758c|M.tuberculosis_H37Rv ---------------------MICFDDVSKVYAHGAT----AVDRLTLEV
MMAR_0034|M.marinum_M MPEAASADQPTTSDAPQSSGLRILLDGVTKRYGVRSTP---AVDNVTLEI
MUL_0033|M.ulcerans_Agy99 MPEAASADQPTTSDAPQSSGLRILLDGVTKRYGVRSTP---AVDNVTLEI
: : : :
Mflv_3158|M.gilvum_PYR-GCK AGGEFVAMLGRSGSGKSTLLRILAGLDNRADGSVLVP--RSR--------
MAV_0140|M.avium_104 EPGAVVALLGASGSGKSTLLRLVAGLDRPSGGRIEIDGKAVRGI-----D
MSMEG_6496|M.smegmatis_MC2_155 SGETFVSVVGPSGSGKTTLLNIVSGIEPHTSGSVRLR-EGSR-------D
MAB_2178|M.abscessus_ATCC_1997 EPGTFLVLVGASGCGKSTLLRLLAGFESPSEGAVRVAGGVPTP------G
Mvan_0123|M.vanbaalenii_PYR-1 DPGSFLVLVGASGCGKSTLLRLLAGFEEPSQGSVQVAGARPTP------G
MLBr_01424|M.leprae_Br4923 DDGEFVVVVGPSGCGKTTSLRMVAGLETLDSGCIRIGDRDVTHVDPK--D
TH_4034|M.thermoresistible__bu ADGEFIILVGPSGCGKSTTLNMIAGLEDITSGEIRIDGERVNEVAPK--D
Mb3784c|M.bovis_AF2122/97 PNGMLTVFVGPSGCGKTTALRMINRMVDPTSGTITVDGTDVSTVNAVKLR
Rv3758c|M.tuberculosis_H37Rv PNGMLTVFVGPSGCGKTTALRMINRMVDPTSGTITVDGTDVSTVNAVKLR
MMAR_0034|M.marinum_M GAGEIVMLVGPSGCGKTTTMKMINRLIEPTSGRIFIGDDDVTRRNPDELR
MUL_0033|M.ulcerans_Agy99 GAGEIVMLVGPSGCGKTTTMKVINRLIEPTSGRIFIGDDDVTRRNPDELR
. .:* **.**:* :.:: : * : :
Mflv_3158|M.gilvum_PYR-GCK ---AVVFQNPRLLPWRRALANVTFALPQAGPDAPSRTARGQ-AALDEVGL
MAV_0140|M.avium_104 PRCAVVFQEPRLLPWRSLAANVAFGLPR-GTERPERTAAVQ-RWLDVVGL
MSMEG_6496|M.smegmatis_MC2_155 ARVGYVFQDPRLLPWRTVMANLGF----VQHERTGCQERAE-HYLDLVGL
MAB_2178|M.abscessus_ATCC_1997 ITSGVVFQQPRLFPWRTVGGNIELALKYAKVPRERWAQRRD-QLLARVGL
Mvan_0123|M.vanbaalenii_PYR-1 VTAGVVFQQPRLFPWRTVGGNVDMALRYAKVPRDKRIERRE-ELLGRVGL
MLBr_01424|M.leprae_Br4923 RDVAMVFQNYALYPHMTVAQNLGFALKISKISKAEIHDRIL-VVAKLLDL
TH_4034|M.thermoresistible__bu RDIAMVFQSYALYPHMTVRQNIAFPLTLAKMKKADIARKVE-ETAKILDL
Mb3784c|M.bovis_AF2122/97 LGIGYVIQNAGLMPHQRVIDNVATVP-VLKGQPRRAARKAGYEVLERVGL
Rv3758c|M.tuberculosis_H37Rv LGIGYVIQNAGLMPHQRVIDNVATVP-VLKGQPRRAARKAGYEVLERVGL
MMAR_0034|M.marinum_M RHIGYVVQGAGLFPHLTVGDNIGIVPRLLKWDKNRVAARID-ELLDLVNL
MUL_0033|M.ulcerans_Agy99 RHIGYVVQGAGVFLHLTVGDNIGIVPRLLKWDKNRVAARID-ELLDLVNL
. *.* : *: :.*
Mflv_3158|M.gilvum_PYR-GCK SG--KARAWPLSLSGGEAQRVSLARALVREPDLLLLDEPFGALDALTRLK
MAV_0140|M.avium_104 RE--FAGHYPRQVSGGMAQRAGLARALARQPSVLLLDEPLAALDALTRLR
MSMEG_6496|M.smegmatis_MC2_155 SH--CADRFPAQLSGGQQQRIGIARAFAVEPDVLLMDEPFSHLDAMTSRT
MAB_2178|M.abscessus_ATCC_1997 EG--TAGRRIWEISGGQQQRVAIARALAAETPLFLLDEPFAALDALTRER
Mvan_0123|M.vanbaalenii_PYR-1 EG--TADRRIWEISGGQQQRVAIARALAAETPLFLLDEPFAALDALTRER
MLBr_01424|M.leprae_Br4923 QP--YLDRKPKDLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRAQ
TH_4034|M.thermoresistible__bu TE--LLDRKPAQLSGGQRQRVAMGRAIVRQPKAFLMDEPLSNLDAKLRVQ
Mb3784c|M.bovis_AF2122/97 DP-KVATRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHE
Rv3758c|M.tuberculosis_H37Rv DP-KVATRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHE
MMAR_0034|M.marinum_M DPAQYRDRFPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQR
MUL_0033|M.ulcerans_Agy99 DPAQYRDRFPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQR
.:*** ** .:.**:. . :*:***:. :*.
Mflv_3158|M.gilvum_PYR-GCK MYRLLHDLWARRRMAVLHVTHDVDEAILLADRVVVLS--DG------AVS
MAV_0140|M.avium_104 MQDLLDAVQQRAGTTTILVTHDVEEAVLLADRVLILRGEEGGAA---TVA
MSMEG_6496|M.smegmatis_MC2_155 LREHLERIWLESRRTVMFVTHDVTEAVQLSDRIVVLAP-GG------YIH
MAB_2178|M.abscessus_ATCC_1997 LQEDVRQVSAESGRTTVFVTHSADEAAFLGSRIVVLTRRPG------QVA
Mvan_0123|M.vanbaalenii_PYR-1 LQEDVRQVSAESGRTTVFVTHSADEAAFLGSRIVVLTRRPG------QVA
MLBr_01424|M.leprae_Br4923 TRNQIAELQCQLGTTTVYVTHDQVEAMTMGDRVAVLCYGVLQQFATPREL
TH_4034|M.thermoresistible__bu MRSEIARLQSRLRTTTVYVTHDQTEAMTLGDRVVVMLDGVAQQIGTPDEL
Mb3784c|M.bovis_AF2122/97 LQNEILRLQAELHKTIVFVTHDIDEALKLADLVAVFAPGGALAQYDETAR
Rv3758c|M.tuberculosis_H37Rv LQNEILRLQAELHKTIVFVTHDIDEALKLADLVAVFAPGGALAQYDETAR
MMAR_0034|M.marinum_M LQDELLRLQEELHKTIVFVTHDFGEAVKLGDRIAILTTGSKVVQYDTPER
MUL_0033|M.ulcerans_Agy99 LQDELLRLQEELHKTIVFVTHDFGEAVKLGDRIAILTTGSKVVQYDTPER
: : . : : ***. ** :.. : ::
Mflv_3158|M.gilvum_PYR-GCK LDRRVDLPFPRSRGDDGFDELRRELLAELGVK---EEVGHDRS-----R-
MAV_0140|M.avium_104 ATHDVAIPKPRDRGDPRIAALREQLLEEVGVPRRGYAIEQEAK-----TS
MSMEG_6496|M.smegmatis_MC2_155 EVIDVELPRPRRASDPAVAVLQAEVLARFEALEKRRTQLQDIV-----D-
MAB_2178|M.abscessus_ATCC_1997 LDIPVDLP--RTGIDP--DDLRRSPEYAQLRTEVGRAVKAAAA-------
Mvan_0123|M.vanbaalenii_PYR-1 LDIPVDLP--RTGVDP--DELRRSPEYGQLRAEVGRAVKAAAA-------
MLBr_01424|M.leprae_Br4923 YHKPENVFVAGFIGSPGMNLVTLSIVDSSVLLGDWPIRIPREI-----AS
TH_4034|M.thermoresistible__bu YTQPANLFVAGFIGSPGMNFFPASLTDAGVRLPFGEALLPPDV-----LD
Mb3784c|M.bovis_AF2122/97 LLSSPANDFVSKFIGLGRGYRWLQLFDAAGLPVRDIEQVSVNG-LSDARD
Rv3758c|M.tuberculosis_H37Rv LLSSPANDFVSKFIGLGRGYRWLQLFDAAGLPVRDIEQVSVNG-LSDARD
MMAR_0034|M.marinum_M ILAEPANDFVRGFVGAGATLMQLSLTRVRDIDLHEAVVAQAGGDPAEAVE
MUL_0033|M.ulcerans_Agy99 VLAEPANDFVRGFVGAGATLMQLSLTRVRDIDLHEAVVAQAGGDPAEAVE
. .
Mflv_3158|M.gilvum_PYR-GCK --------------------------------------------------
MAV_0140|M.avium_104 --------------------------------------------------
MSMEG_6496|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2178|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0123|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01424|M.leprae_Br4923 AAS------EVIIGVRPEHFE----------LGSLGVEVEIDMVEELGAD
TH_4034|M.thermoresistible__bu TVRRHEKTDNIIAGLRPEHLEDVSLLDTYARLGAVTFDVNVELVESLGAE
Mb3784c|M.bovis_AF2122/97 RQ--VRDGWVLVVDGAGAPLGWIDADGRRRHRGGAALSDAMTVGGSVFRP
Rv3758c|M.tuberculosis_H37Rv RQ--VRDGWVLVVDGAGAPLGWIDADGRRRHRGGAALSDAMTVGGSVFRP
MMAR_0034|M.marinum_M LAGSLDREHAIVLDRQNRPLRWLPLEELAKPQALAQVTRDDDL--ECVNL
MUL_0033|M.ulcerans_Agy99 LAGSLDRAHAIVLDRQNRPLRWLPLEELAKPQALAQVTRDDDL--ECVNL
Mflv_3158|M.gilvum_PYR-GCK --------------------------------------------------
MAV_0140|M.avium_104 --------------------------------------------------
MSMEG_6496|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2178|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_0123|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_01424|M.leprae_Br4923 AYLYGRIAGASKVTDQLV-------------VARVDGRNPPAKGSRVRLY
TH_4034|M.thermoresistible__bu KYVHFRAEGAGAHAAQLAQLAAESGAGENEFIARVSTESKAKKGQSLQLA
Mb3784c|M.bovis_AF2122/97 NGNLSQALDAALSSPSGVGVAVDGGGKVIGGILAADVLAEFQKGKKAGGG
Rv3758c|M.tuberculosis_H37Rv NGNLSQALDAALSSPSGVGVAVDGGGKVIGGILAADVLAEFQKGKKAGGG
MMAR_0034|M.marinum_M ASTLNDALDEMLTSSHGV-VVVTGRRNTYQGVIRVETIMEAIADIRGAAN
MUL_0033|M.ulcerans_Agy99 ASTLNDALDEMLTSAHGV-VVVTGRRNTYQGVIRVETIMEAIADIRGAAN
Mflv_3158|M.gilvum_PYR-GCK ------------------------
MAV_0140|M.avium_104 ------------------------
MSMEG_6496|M.smegmatis_MC2_155 ------------------------
MAB_2178|M.abscessus_ATCC_1997 ------------------------
Mvan_0123|M.vanbaalenii_PYR-1 ------------------------
MLBr_01424|M.leprae_Br4923 TEPGNVHFFGVD-GHRIS------
TH_4034|M.thermoresistible__bu FNPAKLLVFDADTGANLTLPPSDA
Mb3784c|M.bovis_AF2122/97 AKPCTT------------------
Rv3758c|M.tuberculosis_H37Rv AKPCTT------------------
MMAR_0034|M.marinum_M SEEPDT------------------
MUL_0033|M.ulcerans_Agy99 SEEPDT------------------