For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KFVGRMRGAAAGLSRRLTVAVAAAAVLPGLVGVVGGSATAGAWSRPGLPVEYLEVPSAAMGRDIRVEFQS GGPGAPALYLLDGMRAREDQNGWDIELPTFEWFLNSGISVVMPVGGQSSFYSDWYKPACGSKDGGCKTYK WETFLTQELPAWLAANRDVKPTGSAVVGLSMAGSSALMLAARHPQQFIYAASLSGTLNPSEGWWPMLIGI SMGDAGGYKADDMWGSTNDPNNAWKANDPTENVATIANNGTRIWVYCGNGKPGELGGTDLPAKFLEGFVC RTNSTFQEKYIEAGGKNGVFNFPQSGTHNWAYWGQQLQAMKPDLQRVLGATPTA*
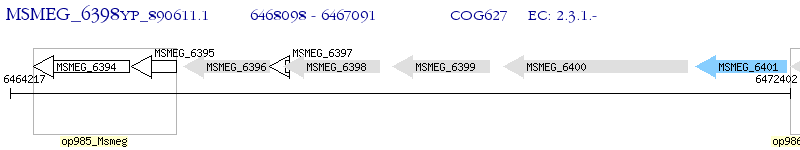
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6398 | - | - | 100% (335) | antigen 85-A |
| M. smegmatis MC2 155 | MSMEG_2078 | - | e-144 | 72.42% (330) | antigen 85-C |
| M. smegmatis MC2 155 | MSMEG_3580 | - | e-124 | 64.95% (331) | antigen 85-C |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3834c | fbpA | 1e-138 | 68.77% (333) | secreted antigen 85-A FBPA (Mycolyl transferase 85A) |
| M. gilvum PYR-GCK | Mflv_1164 | - | 1e-142 | 70.33% (337) | putative esterase |
| M. tuberculosis H37Rv | Rv3804c | fbpA | 1e-138 | 68.77% (333) | secreted antigen 85-A FBPA (Mycolyl transferase 85A) |
| M. leprae Br4923 | MLBr_00097 | fbpA | 1e-131 | 66.57% (335) | antigen 85A, mycolytransferase |
| M. abscessus ATCC 19977 | MAB_0176 | - | 1e-106 | 58.98% (334) | antigen 85-A precursor |
| M. marinum M | MMAR_5368 | fbpA | 1e-134 | 68.47% (333) | secreted antigen 85-A FbpA |
| M. avium 104 | MAV_0214 | - | 1e-138 | 70.15% (335) | antigen 85-A |
| M. thermoresistible (build 8) | TH_2078 | fbpA | 1e-141 | 69.14% (337) | SECRETED ANTIGEN 85-A FBPA (MYCOLYL TRANSFERASE 85A) |
| M. ulcerans Agy99 | MUL_4987 | fbpA | 1e-134 | 68.47% (333) | secreted antigen 85-A FbpA |
| M. vanbaalenii PYR-1 | Mvan_5645 | - | 1e-143 | 70.33% (337) | putative esterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1164|M.gilvum_PYR-GCK MKFVGKL-----RAAARRSTVVALAAIVLPGLISAVGG---SATAGAFSR
Mvan_5645|M.vanbaalenii_PYR-1 MKLVGALPRV-IRSMGRRLTVAALAAIVLPGLISAVGG---SATAGAFSR
TH_2078|M.thermoresistible__bu MKLVAKLRGA-ARALPRRLTVAALAAALLPGLVGAFGG---SATAGAFSR
MSMEG_6398|M.smegmatis_MC2_155 MKFVGRMRGA-AAGLSRRLTVAVAAAAVLPGLVGVVGG---SATAGAWSR
MMAR_5368|M.marinum_M MKLVDRFRGA-VTGTPRRLMVGAVGAALLSGLVGFVGG---SATASAFSR
MUL_4987|M.ulcerans_Agy99 MKLVDRFRGA-ATGTSRRLMVGAVGAALLSGLVGFVGG---SATASAFSR
MAV_0214|M.avium_104 MTLVDRLRGA-VAGMPRRLVVGAAGAALLSGLIGAVGG---SATAGAFSR
Mb3834c|M.bovis_AF2122/97 MQLVDRVRGA-VTGMSRRLVVGAVGAALVSGLVGAVGG---TATAGAFSR
Rv3804c|M.tuberculosis_H37Rv MQLVDRVRGA-VTGMSRRLVVGAVGAALVSGLVGAVGG---TATAGAFSR
MLBr_00097|M.leprae_Br4923 MKFVDRFRGA-VAGMLRRLVVEAMGVALLSALIGVVG----SAPAEAFSR
MAB_0176|M.abscessus_ATCC_1997 MKLFSKMRGALARQSARRIAVAATAVAVLPGVAGIVGGTALTPVAGAFSR
* :. . ** * . .. ::..: . .* :. * *:**
Mflv_1164|M.gilvum_PYR-GCK PGLPVEYLMVPSAGMGRDIKVQFQSGGPNAPGVYLLDGLRAQDDFNGWDI
Mvan_5645|M.vanbaalenii_PYR-1 PGLPVEYLMVPSAGMGRDIKVQFQSGGPNAPGVYLLDGLRAQDDFSGWDI
TH_2078|M.thermoresistible__bu PGLPVEYLMVPSAAMGRDIKVQFQSGGEGSPGVYLLDGLRARDDFNGWDI
MSMEG_6398|M.smegmatis_MC2_155 PGLPVEYLEVPSAAMGRDIRVEFQSGGPGAPALYLLDGMRAREDQNGWDI
MMAR_5368|M.marinum_M PGLPVEYLQVPSAAMGRNIKVQFQSGGANSPALYLLDGMRAQDDFSGWDI
MUL_4987|M.ulcerans_Agy99 PGLPVEYLQVPSVAMGRNIKVQFQSGGANSPALYLLDGMRAQDDFSGWDI
MAV_0214|M.avium_104 PGLPVEYLQVPSAAMGRDIKVQFQSGGANSPALYLLDGMRAQDDFNGWDI
Mb3834c|M.bovis_AF2122/97 PGLPVEYLQVPSPSMGRDIKVQFQSGGANSPALYLLDGLRAQDDFSGWDI
Rv3804c|M.tuberculosis_H37Rv PGLPVEYLQVPSPSMGRDIKVQFQSGGANSPALYLLDGLRAQDDFSGWDI
MLBr_00097|M.leprae_Br4923 PGLPVEYLQVPSPSMGRDIKVQFQNGGANSPALYLLDGLRAQDDFSGWDI
MAB_0176|M.abscessus_ATCC_1997 PGLPVEYLQVPSASMGREIKVQFQPGG--AKAVYLLDGLRARDDFSGWDI
******** *** .***:*:*:** ** : .:*****:**::* .****
Mflv_1164|M.gilvum_PYR-GCK NTPAFEWYLDSGLAVVMPVGGQSSFYSDWYSPACG--KAGCSTYKWETFL
Mvan_5645|M.vanbaalenii_PYR-1 NTQAFEWYLDSGLAVVMPVGGQSSFYSDWYAPACG--KAGCSTYKWETFL
TH_2078|M.thermoresistible__bu ETAAFEWYYESGLSVIMPVGGQSSFYSDWYSPACG--NNGCQTYKWETFL
MSMEG_6398|M.smegmatis_MC2_155 ELPTFEWFLNSGISVVMPVGGQSSFYSDWYKPACGSKDGGCKTYKWETFL
MMAR_5368|M.marinum_M NTPAFEWYYQSGISVAMPVGGQSSFYSDWYNPACG--KAGCTTYKWETFL
MUL_4987|M.ulcerans_Agy99 NTPAFEWYYQSGISVAMPVGGQSSFYSDWYNPACG--KAGCTTYKWETFL
MAV_0214|M.avium_104 NTPAFEWYNQSGISVAMPVGGQSSFYSDWYKPACG--KAGCTTYKWETFL
Mb3834c|M.bovis_AF2122/97 NTPAFEWYDQSGLSVVMPVGGQSSFYSDWYQPACG--KAGCQTYKWETFL
Rv3804c|M.tuberculosis_H37Rv NTPAFEWYDQSGLSVVMPVGGQSSFYSDWYQPACG--KAGCQTYKWETFL
MLBr_00097|M.leprae_Br4923 NTTAFEWYYQSGISVVMPVGGQSSFYSDWYSPACG--KAGCQTYKWETFL
MAB_0176|M.abscessus_ATCC_1997 ETTAFEDYYQSGISMVMPVGGQSSFYTDWYNPAKG--KDGVWTYKWETFT
: :** : :**::: **********:*** ** * . * *******
Mflv_1164|M.gilvum_PYR-GCK TQELPAYLAANKGVDPNRNAAIGLSMAGSAALTLAIYYPQQFQYAGSMSG
Mvan_5645|M.vanbaalenii_PYR-1 TQELPAYLAANKGVDPNRNAAVGLSMAGSAALTLAIYHPQQFQYAGSLSG
TH_2078|M.thermoresistible__bu TQELPAYLASNYGVDRNRNAAVGLSMAGSAALVLAIYYPQQFQYAASLSG
MSMEG_6398|M.smegmatis_MC2_155 TQELPAWLAANRDVKPTGSAVVGLSMAGSSALMLAARHPQQFIYAASLSG
MMAR_5368|M.marinum_M TSELPQYLSANKGVKPTGSGVVGLSMAGSSALILAAYHPDQFVYSGSLSA
MUL_4987|M.ulcerans_Agy99 TSELPQYLSANKGVKPTGSGVVGLSMAGSSALILAAYHPDQFVYSGSLSA
MAV_0214|M.avium_104 TSELPQYLSAQKQVKPTGSGVVGLSMAGSSALILAAYHPDQFVYAGSLSA
Mb3834c|M.bovis_AF2122/97 TSELPGWLQANRHVKPTGSAVVGLSMAASSALTLAIYHPQQFVYAGAMSG
Rv3804c|M.tuberculosis_H37Rv TSELPGWLQANRHVKPTGSAVVGLSMAASSALTLAIYHPQQFVYAGAMSG
MLBr_00097|M.leprae_Br4923 TSELPQYLQSNKQIKPTGSAAVGLSMAGLSALTLAIYHPDQFIYVGSMSG
MAB_0176|M.abscessus_ATCC_1997 TQELPAYLAANKGISQTGNAVVGLSMGASAALTLSIYHPQLFVYAGALSG
*.*** :* :: :. . ...:****.. :** *: :*: * * .::*.
Mflv_1164|M.gilvum_PYR-GCK FLNLSEGWWPALVNISMGDAGGYDANDMWGRTEDPNSAWKRNDPMVNIGK
Mvan_5645|M.vanbaalenii_PYR-1 YLNLSEGWWPMLVNISMGDAGGYDANDMWGRTEDPDSAWKRNDPMVNIAK
TH_2078|M.thermoresistible__bu FLNLSEGWWPMLVGLAMGDAGGYDPNDMWGRTEDPNSAWKRNDPMVNIDK
MSMEG_6398|M.smegmatis_MC2_155 TLNPSEGWWPMLIGISMGDAGGYKADDMWGSTNDPNNAWKANDPTENVAT
MMAR_5368|M.marinum_M LLDPSQGMGPSLIGLAMGDAGGYKASDMWGPKDDP--AWARNDPMLQVGK
MUL_4987|M.ulcerans_Agy99 LLDPSQGIGPSLIGLAMGDAGGYKASDMWGPKDDP--AWARNDPMLQVGK
MAV_0214|M.avium_104 LLDPSQGMGPSLIGLAMGDAGGYKAADMWGPKEDP--AWARNDPSLQVGK
Mb3834c|M.bovis_AF2122/97 LLDPSQAMGPTLIGLAMGDAGGYKASDMWGPKEDP--AWQRNDPLLNVGK
Rv3804c|M.tuberculosis_H37Rv LLDPSQAMGPTLIGLAMGDAGGYKASDMWGPKEDP--AWQRNDPLLNVGK
MLBr_00097|M.leprae_Br4923 LLDPSNAMGPSLIGLAMGDAGGYKAADMWGPSTDP--AWKRNDPTVNVGT
MAB_0176|M.abscessus_ATCC_1997 FLNPSD--MKFQIGLAMGDAGGFSASDMWGPDSDP--AWQRNDPFLNIQK
*: *: :.::******:.. **** ** ** *** :: .
Mflv_1164|M.gilvum_PYR-GCK LVANNTRIWVYCGDGKPAEVNGNVA-GNNLPAKFLEGLTIRTNRTFQEEY
Mvan_5645|M.vanbaalenii_PYR-1 LVANNTRIWVFCGNGKPAEVNGNVA-GNNLPAKFLEGFTLRTNKTFQEEY
TH_2078|M.thermoresistible__bu LVANNTRIWVFCGDGTPADIGAGVPEGDNFNAKFLESFTLRTNKTFQQEY
MSMEG_6398|M.smegmatis_MC2_155 IANNGTRIWVYCGNGKPGELG-----GTDLPAKFLEGFVCRTNSTFQEKY
MMAR_5368|M.marinum_M LVANNTRIWVYCGNGKPSDLG-----GDNLPAKFLEGFVRTSNMKFQAAY
MUL_4987|M.ulcerans_Agy99 LVANNTRIWVYCGNGKPSDLG-----GDNLPAKFLEGFVRTSNMKFQAAY
MAV_0214|M.avium_104 LVANNTRIWVYCGNGKPSDLG-----GDNLPAKFLEGFVRTSNLKFQDAY
Mb3834c|M.bovis_AF2122/97 LIANNTRVWVYCGNGKPSDLG-----GNNLPAKFLEGFVRTSNIKFQDAY
Rv3804c|M.tuberculosis_H37Rv LIANNTRVWVYCGNGKPSDLG-----GNNLPAKFLEGFVRTSNIKFQDAY
MLBr_00097|M.leprae_Br4923 LIANNTRIWMYCGNGKPTELG-----GNNLPAKLLEGLVRTSNIKFQDGY
MAB_0176|M.abscessus_ATCC_1997 IIDNGTRLWIYCGTGDSTDLDATRNGFENFTGGFLEGMAIGSNKKFVEAY
: *.**:*::** * . ::. :: . :**.:. :* .* *
Mflv_1164|M.gilvum_PYR-GCK IAAGGKNGVFNFPANGTHDWPYWGQQLQQMKPDIQRVLGAVPQPSAPAAP
Mvan_5645|M.vanbaalenii_PYR-1 LAAGGKNGVFNFPSGGTHDWPYWGQQLQQMKPDIQRVLGAVPQPSAPAAP
TH_2078|M.thermoresistible__bu IAAGGKNGVFNFPDAGTHSWGYWGQQLQQMKPDIQRVLGVS------AA-
MSMEG_6398|M.smegmatis_MC2_155 IEAGGKNGVFNFPQSGTHNWAYWGQQLQAMKPDLQRVLGATP-----TA-
MMAR_5368|M.marinum_M NAAGGHNAVWNFDDNGTHSWEYWGAQLNAMKPDLQHTLGATP--------
MUL_4987|M.ulcerans_Agy99 NAAGGHNAVWNFDDNGTHSWEYWGAQLNAMRPDLQHTLGATP--------
MAV_0214|M.avium_104 NGAGGHNAVWNFDANGTHDWPYWGAQLQAMKPDLQSVLGATPGAGPATAA
Mb3834c|M.bovis_AF2122/97 NAGGGHNGVFDFPDSGTHSWEYWGAQLNAMKPDLQRALGATPN-------
Rv3804c|M.tuberculosis_H37Rv NAGGGHNGVFDFPDSGTHSWEYWGAQLNAMKPDLQRALGATPN-------
MLBr_00097|M.leprae_Br4923 NAGGGHNAVFNFPDSGTHSWEYWGEQLNDMKPDLQQYLGATP--------
MAB_0176|M.abscessus_ATCC_1997 TAAGGKNAHIEFPPGGIHNWTYWGQQLRAMKPDMVAYLQSH---------
.**:*. :* * *.* *** **. *:**: *
Mflv_1164|M.gilvum_PYR-GCK VAAEAPGAGG
Mvan_5645|M.vanbaalenii_PYR-1 VDAVAPTSGG
TH_2078|M.thermoresistible__bu ----------
MSMEG_6398|M.smegmatis_MC2_155 ----------
MMAR_5368|M.marinum_M --NTGDTQGA
MUL_4987|M.ulcerans_Agy99 --NTGDTQGA
MAV_0214|M.avium_104 ATNAGNGQGT
Mb3834c|M.bovis_AF2122/97 --TGPAPQGA
Rv3804c|M.tuberculosis_H37Rv --TGPAPQGA
MLBr_00097|M.leprae_Br4923 --------GA
MAB_0176|M.abscessus_ATCC_1997 ----------