For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGGVYAPVPPGVPAPTPTPVKPRRRILRRLLITSLGLATVLAVAFVAFVFWPNPTVPADQPNMTQVMTG TDNDWLSFDDFVPGTAIAADGSDNDEYECTLGFLGRDSSGTPIGFTAGHCDRPGDDSPHFTRKADEDTPV ELGRFTTVQTADMPDKVVKTGPDGWSDFAVIEIDPSLTDSAEDNARIAGVYRVTKVLDYEYLLNNNVTLC KFGFVSGETCGKVIGHDNLTVEAELYSASGDSGSPAYVKFGNDEVAAVGILRGSPEGEDNITEFSLLAPI LEATSSSLFCDDPTCRGNGS
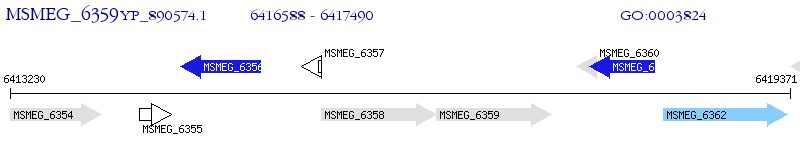
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6359 | - | - | 100% (300) | trypsin domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_6180 | - | 3e-07 | 30.77% (169) | secreted protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3692c | - | 9e-07 | 26.52% (181) | putative protease |
| M. gilvum PYR-GCK | Mflv_0488 | - | 4e-13 | 30.30% (198) | hypothetical protein Mflv_0488 |
| M. tuberculosis H37Rv | Rv3668c | - | 9e-07 | 26.52% (181) | protease |
| M. leprae Br4923 | MLBr_02242 | - | 5e-17 | 34.39% (189) | hypothetical protein MLBr_02242 |
| M. abscessus ATCC 19977 | MAB_3348 | - | 9e-09 | 26.67% (180) | hypothetical protein MAB_3348 |
| M. marinum M | MMAR_2300 | - | 2e-10 | 28.63% (255) | hypothetical protein MMAR_2300 |
| M. avium 104 | MAV_0678 | - | 1e-22 | 35.42% (192) | hypothetical protein MAV_0678 |
| M. thermoresistible (build 8) | TH_0965 | - | 3e-06 | 28.57% (175) | POSSIBLE PROTEASE |
| M. ulcerans Agy99 | MUL_1503 | - | 6e-11 | 29.02% (255) | hypothetical protein MUL_1503 |
| M. vanbaalenii PYR-1 | Mvan_3169 | - | 1e-08 | 29.07% (172) | hypothetical protein Mvan_3169 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02242|M.leprae_Br4923 --------------------------------------------------
MAV_0678|M.avium_104 --------------------------------------------------
MMAR_2300|M.marinum_M MVGVSAHALANLDSPSSLARTALLLAGALLCLSVAAWPVLSQRRRQHPET
MUL_1503|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0488|M.gilvum_PYR-GCK --------------------------------------------------
Mb3692c|M.bovis_AF2122/97 --------------------------------------------------
Rv3668c|M.tuberculosis_H37Rv --------------------------------------------------
TH_0965|M.thermoresistible__bu --------------------------------------------------
MAB_3348|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3169|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6359|M.smegmatis_MC2_155 -------------------------------------------------M
MLBr_02242|M.leprae_Br4923 -------------------------MRSIYWFTGETCATVRWLRGLGLAS
MAV_0678|M.avium_104 --------------------------MTVSWLRGSGLVGAVLVAATAVAK
MMAR_2300|M.marinum_M QEQLFAGTNVPYAAPPQRRQSDGGRLTIAALICAAGFVAAIWLPSKSTMP
MUL_1503|M.ulcerans_Agy99 ---MFAGTNVPYAAPPQRRQSDGGRLTIAALICAAGFVAAVWLPSKSTMP
Mflv_0488|M.gilvum_PYR-GCK --------------------------MMRNLLKTVGGTAAAALAALTFTQ
Mb3692c|M.bovis_AF2122/97 ------------------------------MQTAHRRFAAAFAAVLLAVV
Rv3668c|M.tuberculosis_H37Rv ------------------------------MQTAHRRFAAAFAAVLLAVV
TH_0965|M.thermoresistible__bu ------------------------------------MLPIVALTGLLVAA
MAB_3348|M.abscessus_ATCC_1997 -------------------MTSVGDKGRRTGPWGRLTAYAAAVAALSGIG
Mvan_3169|M.vanbaalenii_PYR-1 -------------------MS-----GRSSS--RVLHLLFAVLAPVLALA
MSMEG_6359|M.smegmatis_MC2_155 TGGVYAPVPPGVPAPTPTPVKPRRRILRRLLITSLGLATVLAVAFVAFVF
MLBr_02242|M.leprae_Br4923 AAVIAPKVFASMVLAEYA---------------PSPGLRLCDES-SICTA
MAV_0678|M.avium_104 TAGAAP----PPALPAPP---------------PSPGIGVDHEN-GRCTA
MMAR_2300|M.marinum_M ARPLVALPGASAIAPGTG---------------IAVTYADGSGG-MGCTA
MUL_1503|M.ulcerans_Agy99 ARPLVALPGASAIAPGTG---------------IAVTYADGSGG-MGCTA
Mflv_0488|M.gilvum_PYR-GCK APVAHAFGEYPVASPGAH---------------LVSTTLAGNGLPYQCTA
Mb3692c|M.bovis_AF2122/97 CLPANTAAADDKLPLGGG---------------AGIVVNG-DTMCTLTTI
Rv3668c|M.tuberculosis_H37Rv CLPANTAAADDKLPLGGG---------------AGIVVNG-DTMCTLTTI
TH_0965|M.thermoresistible__bu LVPSVVAHAAGPVPLGGG---------------SGLVVNG-ETLCTLTTI
MAB_3348|M.abscessus_ATCC_1997 LANPIAAHADP--LIYPG---------------MEIIQRPPGKESLSCTL
Mvan_3169|M.vanbaalenii_PYR-1 LQT-VPAQAQPGVLVFPG---------------MEIHQ-----DTVLCTL
MSMEG_6359|M.smegmatis_MC2_155 WPNPTVPADQPNMTQVMTGTDNDWLSFDDFVPGTAIAADGSDNDEYECTL
*
MLBr_02242|M.leprae_Br4923 ACVARGNDGSSYLLTIGHCDAYDGSVWPYGPDVPLGK-------RTVSEN
MAV_0678|M.avium_104 GFAAQGSDGSYYLMTSGHCDAHDGAEWTYGNDAPLGR-------ISASEH
MMAR_2300|M.marinum_M GFLVRTNAGRTGILTAGHCNK-EGEASKVSINYSAGG-------GYVNIG
MUL_1503|M.ulcerans_Agy99 GFLVRTNAGRTGILTAGHCNK-EGEASKVSINYSTGG-------GYVNIG
Mflv_0488|M.gilvum_PYR-GCK GFTVRTPGGSPGMLTAGHCDR-DPLNDTVLQRTPTGD-------RVVGRY
Mb3692c|M.bovis_AF2122/97 GHDKNGDLIG---FTSAHCGGPGAQIAAEG----------------AENA
Rv3668c|M.tuberculosis_H37Rv GHDKNGDLIG---FTSAHCGGPGAQIAAEG----------------AENA
TH_0965|M.thermoresistible__bu GHDNRGALVG---FTSAHCGGPGAVVSAEG----------------AEDA
MAB_3348|M.abscessus_ATCC_1997 GFVDPAARVG---VTAGHCGGNGPVFTSDGVRIGQLA-------VAQSNI
Mvan_3169|M.vanbaalenii_PYR-1 GYVDPQTRIG---YTAGHCRASGTVSDKFGTMIGTQG-------TFRDNT
MSMEG_6359|M.smegmatis_MC2_155 GFLGRDSSGTPIGFTAGHCDRPGDDSPHFTRKADEDTPVELGRFTTVQTA
. * .**
MLBr_02242|M.leprae_Br4923 EGDR----------KDAAIILLDLSQGYSSGD----VYS-QLLRNVLSEN
MAV_0678|M.avium_104 EGDK----------RDAAIIRLEPSVGMPVGD----VGGRYQVRDVLSRP
MMAR_2300|M.marinum_M TFSQSVSEGLNGEAHDIGLITLDSGKIPQSPA----IKAAVPVTGIATD-
MUL_1503|M.ulcerans_Agy99 TFSQSVSEGLNGEAHDIGLITLDSGKIPQSPA----IKAAVPVTGIATD-
Mflv_0488|M.gilvum_PYR-GCK VRWEVLPG-----VRDVGLVDLAGSSVPLVDQ----VDGMRVSRVMSGDD
Mb3692c|M.bovis_AF2122/97 GPVGIMVAG--NDGLDYAVIKFDPAKVTPVAV----FNGFAIN-GIGPD-
Rv3668c|M.tuberculosis_H37Rv GPVGIMVAG--NDGLDYAVIKFDPAKVTPVAV----FNGFAIN-GIGPD-
TH_0965|M.thermoresistible__bu GVLGTMVAG--NDALDYAVIEFDPQKVIPVNE----VNGFRID-GLGPD-
MAB_3348|M.abscessus_ATCC_1997 KPEGPLARD--DYRIDYEGIAFDDGVPINNVL----PNGLRLGVDPAVV-
Mvan_3169|M.vanbaalenii_PYR-1 PDGMTVDTN--HQIIDWEVIHLAPHVAVNNVL----PGGKVLVTDPAVL-
MSMEG_6359|M.smegmatis_MC2_155 DMPDKVVKTGPDGWSDFAVIEIDPSLTDSAEDNARIAGVYRVTKVLDYEY
* : :
MLBr_02242|M.leprae_Br4923 EIQLGMLFCKVGAITG-ETCGTIKSMD--GDVVEASVYSLSAGSGSPGFV
MAV_0678|M.avium_104 QIQVGMPFCKIGAVTG-ETCGAIKGVD--GDVVEASVFSLDGDSGSPGFV
MMAR_2300|M.marinum_M -LKVGQLLCKFGMKTGRAECGQVTDIS--ASKVAFLAASECGDSGGPVYR
MUL_1503|M.ulcerans_Agy99 -LKVGQLLCKFGMKTGRAECGQVTDIS--ASKVAFLAASECGDSGGPVYR
Mflv_0488|M.gilvum_PYR-GCK VRREQPELCKSGARTG-LSCGPVTSVT--DNQVSFRAWDDLGDSGAPVYA
Mb3692c|M.bovis_AF2122/97 -PSFGQIACKQGRTTG-NSCGVTWGPGESPGTLVMQVCGGPGDSGAPVTV
Rv3668c|M.tuberculosis_H37Rv -PSFGQIACKQGRTTG-NSCGVTWGPGESPGTLVMQVCGGPGDSGAPVTV
TH_0965|M.thermoresistible__bu -PVFGEIACKLGRTTG-HSCGVTWGPGQQPGTIVNQVCGQPGDSGAPVTV
MAB_3348|M.abscessus_ATCC_1997 -PRDGLPVCRVGITTG-EACGAIAGFGNGWF-LVDGVPAEHGDSGSAVYT
Mvan_3169|M.vanbaalenii_PYR-1 -PVRGMPVCHFGVVTG-ESCGTIESVNNGWFTMANGVVSRKGDSGGPVYT
MSMEG_6359|M.smegmatis_MC2_155 LLNNNVTLCKFGFVSG-ETCGKVIGHDN--LTVEAELYSASGDSGSPAYV
*: * :* ** . : ..**..
MLBr_02242|M.leprae_Br4923 KNVDGIVSAVGILMSSPEGDDHTT---------YFVPMPPLLEKRGLQSF
MAV_0678|M.avium_104 MNPDGSVSAVGLLMSSPDGDDYTT---------YFMLVNPLLDRWGLRIL
MMAR_2300|M.marinum_M LDDDGTAVAVGILIRAGHANDPHPGCSTPATFSIAELVRPWLDRWSLTAI
MUL_1503|M.ulcerans_Agy99 LDDDGTAVAVGILIRAGHANDPHPGCSTPATFSIAELVRPWLDRWSLTAI
Mflv_0488|M.gilvum_PYR-GCK RQPDGTVAAVGILF--AHSDDAFG------RIVHASLVAPVMSLWGLSTW
Mb3692c|M.bovis_AF2122/97 DN-----LLVGMIHGAFSDNLPSCITKYIPLHTPAVVMSINADLADINAK
Rv3668c|M.tuberculosis_H37Rv DN-----LLVGMIHGAFSDNLPSCITKYIPLHTPAVVMSINADLADINAK
TH_0965|M.thermoresistible__bu NN-----RLVGMIHGAFSEVLPTCVVKFIPLHTPAVTISFNTQLADINAK
MAB_3348|M.abscessus_ATCC_1997 VTSPGNAAIVGIVSARFTANQTRRESTVALSWPTVQQQIRNDIAASRRPE
Mvan_3169|M.vanbaalenii_PYR-1 PTPDGRTVLVGLFNS------TWGTLPAAISWQVASQMAEADTIS-----
MSMEG_6359|M.smegmatis_MC2_155 KFGNDEVAAVGILRGSPEGEDNIT---------EFSLLAPILEATSSSLF
**:.
MLBr_02242|M.leprae_Br4923 PEVLKRLSPRLVGWGSGS-------------------------
MAV_0678|M.avium_104 P------------------------------------------
MMAR_2300|M.marinum_M TEPGSP-------------------------------------
MUL_1503|M.ulcerans_Agy99 TEPGSP-------------------------------------
Mflv_0488|M.gilvum_PYR-GCK G------------------------------------------
Mb3692c|M.bovis_AF2122/97 NRPGAGFVPVPA-------------------------------
Rv3668c|M.tuberculosis_H37Rv NRPGAGFVPVPA-------------------------------
TH_0965|M.thermoresistible__bu NRPGAGFVPIGAVP-----------------------------
MAB_3348|M.abscessus_ATCC_1997 IASPVASNPQPPPPAINPQAPPMVLPPAPSDDRPSGPARMVVP
Mvan_3169|M.vanbaalenii_PYR-1 -AASAVVAPQP--------------------------------
MSMEG_6359|M.smegmatis_MC2_155 CDDPTCRGNGS--------------------------------