For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LNLKSFVAAAQRFWSTYLLVAGLVLIVGAAAIMMLPVTYVSSARLMVSIEGSTTAAAYQNEEVAIRRIRT YIPLLKSDVVTQRVIDKLGLPMTAKQLAEELSATNVPPKTSLIDIRISDSSPQRAELIANTVAREFVAYT AAIETATGEDDQKVQTTMVSDATPARRDPVERALLYLLAALAALLLGGAAVWLRASREQLDADSDDPVGR APKAAQTVEGS
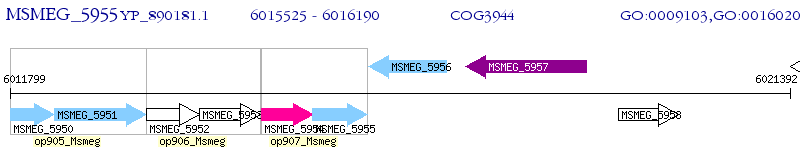
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5955 | - | - | 100% (221) | protein-tyrosine kinase, putative |
| M. smegmatis MC2 155 | MSMEG_5954 | - | 1e-09 | 26.53% (196) | cell surface polysaccharide biosynthesis, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4771 | - | 1e-21 | 33.33% (192) | lipopolysaccharide biosynthesis |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4516 | - | 9e-12 | 28.70% (230) | hypothetical protein MMAR_4516 |
| M. avium 104 | MAV_1135 | - | 1e-14 | 32.22% (180) | chain length determinant protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1681 | - | 1e-22 | 42.24% (161) | lipopolysaccharide biosynthesis |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4771|M.gilvum_PYR-GCK MNLQDFVKLLRSRWMTVCVTMVFAVMGAIAYTLLITPQYQASTRLFVSTS
Mvan_1681|M.vanbaalenii_PYR-1 MNLQDFVKLLRARWITLCVALIVAVLGAVAVSMLTTPLYQASTRLFVSTA
MSMEG_5955|M.smegmatis_MC2_155 MNLKSFVAAAQRFWSTYLLVAGLVLIVGAAAIMMLPVTYVSSARLMVSIE
MMAR_4516|M.marinum_M MDFRTFVQTLAKRWKLVVGALLACLAGATAVTAFQTKVYQSSATIFISVS
MAV_1135|M.avium_104 MDFRTFVRILGAHWKLALTALLACTVGAAFVTALQTKHYQSSATVLISFS
*::: ** * . . : . * :*: :::*
Mflv_4771|M.gilvum_PYR-GCK SSSSVSEIYQGNRFSQERVVSYAELLMGETLAQRTIDKLGLDMSASSLRE
Mvan_1681|M.vanbaalenii_PYR-1 AGSSLSDMYQGNRFSQERVVSYAELLKGETLAQRTIDKLGLDMTAGALRA
MSMEG_5955|M.smegmatis_MC2_155 G-STTAAAYQNEEVAIRRIRTYIPLLKSDVVTQRVIDKLGLPMTAKQLAE
MMAR_4516|M.marinum_M GQTDVNDVYWGGQAAQDLLSSYAEIAGGREVAQRAISQLQLPISPEALVQ
MAV_1135|M.avium_104 GATDLNEVYSGTETAQERLSSYAQIAGGHIVAERAIRQLQLPMSADDLLS
. : * . . : : :* : . :::*.* :* * ::. *
Mflv_4771|M.gilvum_PYR-GCK NISAS-AKPDTVLIDVAVRDPSPVRARDIANTLSDEFVAMVRELETPDGG
Mvan_1681|M.vanbaalenii_PYR-1 NVSAS-AKPDTVLIDVKVLDPSPVRARDIANTLSDEFVAMIRELETPEDG
MSMEG_5955|M.smegmatis_MC2_155 ELSATNVPPKTSLIDIRISDSSPQRAELIANTVAREFVAYTAAIETATGE
MMAR_4516|M.marinum_M QTQVK-YTPKSMLFTIAVSDTDPKRAALLAGGMADSFATLVGTLG--SAP
MAV_1135|M.avium_104 QTQVK-YTPKSVMFTISVKDTDPARAAALAGAMANQFSALLPTIAPNTGP
: ... *.: :: : : *..* ** :*. :: .* : :
Mflv_4771|M.gilvum_PYR-GCK ATPDAR-------------------------------------VVVEQRA
Mvan_1681|M.vanbaalenii_PYR-1 SRPDSR-------------------------------------VVVEQRA
MSMEG_5955|M.smegmatis_MC2_155 DDQKVQ-------------------------------------TTMVSDA
MMAR_4516|M.marinum_M RALGAE-------------PQQQAPPTAP-----------MARVTVVKRA
MAV_1135|M.avium_104 NTVGAQATAGPRPEVAETTPAPGQPPEAPAAVGPTGKPWPLARATMVEPP
. ..: . .
Mflv_4771|M.gilvum_PYR-GCK TIPEHPVVPNTTRNIAMGLALGVLLGVAIAVVRDLLDNTVKDRETLEGIT
Mvan_1681|M.vanbaalenii_PYR-1 SLPVTPVVPKTGRNIMFGVALGLLLGFGVAIVRDLLDNTVKDRKTLEEIA
MSMEG_5955|M.smegmatis_MC2_155 TPARRDPVERALLYLLAALAALLLGGAAVWLR--------ASREQLDADS
MMAR_4516|M.marinum_M EVPVSPSRPLPKRNLVMGLAAGLLLGIAVALAREASDRTVHSRRQLDQLS
MAV_1135|M.avium_104 RVPNVPVSPVPLRNMVIGFIAGLLLAIAVALTREAGDRTVRTRAKLEELS
. . : .. :* . .: : * *: :
Mflv_4771|M.gilvum_PYR-GCK GAGIVGSVPLDKERRKQAAVAFDRDNSGIAEAFRKLRTNLQFLAVDNPPR
Mvan_1681|M.vanbaalenii_PYR-1 GTGVVGAVPLDKERRKQAAISFDSDNSGISEAFRKLRTNLQFLAVDHPPR
MSMEG_5955|M.smegmatis_MC2_155 DDPVG--------RAPKAAQTVEGS-------------------------
MMAR_4516|M.marinum_M GLPTLAELPGRRGGVPQFGNDAAIDDAVRGLRTRLLR------AMGPQAR
MAV_1135|M.avium_104 GLPTLAELPGRRGGTPRFGTDTAFDDAVRGMRTRLLR------AMGPDAR
. : . .
Mflv_4771|M.gilvum_PYR-GCK VIVVTSSMPSEGKSTTAINIALALAEGEHSVVLVDGDMRRPSLHKYLGLV
Mvan_1681|M.vanbaalenii_PYR-1 VIVVTSSMPGEGKSTTAINIALALAEAGRNVALIDGDLRRPTLHKYLDLV
MSMEG_5955|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4516|M.marinum_M RLVVAAPFAGEGTTTTALNLSLSLAELGEDVLLVEGDTRRPVIAGLLKVE
MAV_1135|M.avium_104 QVLVTGPFGGEGTTTTAMNLALALAEMGEDALLVEGDSRRHVIAGLLRVE
Mflv_4771|M.gilvum_PYR-GCK GPVGFSTVLSGGTSLSEALQKTRFPGLTVLTSGAVPPN--PSELLGSQSA
Mvan_1681|M.vanbaalenii_PYR-1 GAVGFSTVLSGGVGLADALQKTRFPGLTVLTSGTIPPN--PSELLGSLAA
MSMEG_5955|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4516|M.marinum_M SREGLANALANPEIALEVVKPTRISKLFILASRSVRRETASANAYLPEVI
MAV_1135|M.avium_104 SGEGLASALANPAAAAEVVKPTPISRLFILASRSARRESLPPSAYLPEVV
Mflv_4771|M.gilvum_PYR-GCK RKLLSELRAEFDYVIVDSTPLLAVTDAAILGAGADGVLVMARFGQTKRDQ
Mvan_1681|M.vanbaalenii_PYR-1 RKVVNELRAQFDYVVIDSTPLVAVTDAAILAAGADGVLIIARYGHTKREQ
MSMEG_5955|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4516|M.marinum_M DSVLMDLSARFSRMVIDGPPVLATADTGLLAGAVQATVLVVRAGRTTVDE
MAV_1135|M.avium_104 DRVVQELSSRFDRIVIDGPPVLATADTGLLSGAVPATVLVVRAKRTTADE
Mflv_4771|M.gilvum_PYR-GCK LAHAVGSLVDVGAPLLGAVFTMVPTRGSSSYSYSYSYYGSESAPQQARQD
Mvan_1681|M.vanbaalenii_PYR-1 LAHAVGSLEGVGAPLLGAILTMMPTRGTGSYGYSYSYYGDDSTSSSSSR-
MSMEG_5955|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4516|M.marinum_M VDDALSALRAAGANVVGTVLTDARISRHTSAAARIYRGKISGLS------
MAV_1135|M.avium_104 LRDALTALRAAGAHVVGTVLTDARQSLHIRAADRAYRSKVSRPA------
Mflv_4771|M.gilvum_PYR-GCK VPVAPTKPAALDAAPSQTGAEPPDPDSSLPRADATPGASRRRRRQVSE
Mvan_1681|M.vanbaalenii_PYR-1 --RMSAKLAALELPPPKSDHSLKPPQSIAESGPDIVQQTSAKRRRAAD
MSMEG_5955|M.smegmatis_MC2_155 ------------------------------------------------
MMAR_4516|M.marinum_M ------------------------------------------------
MAV_1135|M.avium_104 ------------------------------------------------