For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPLQVIDIDDPADPRIDDFRDLNSVDRRPDLPTGKALVIAEGVLVVQRMLASRFIPRALLGTDRRLGELA ADLDRYPQYADVPFYRASAEVMAEAIGFHLNRGVLASAPRAAELTVDDVIADARTIAVLEGVNDHENLGS IFRNAAGLAVDAVIFGAGCADPLYRRAVRVSMGHALLVPFARASSWPTDLELLRDKGFRLLAMTPDPSAA TLSDAMVQLEGDRVAILVGAEGPGLTERTMRASDVRVRIPMSRGTDSLNVATAAALAFYERARLSH*
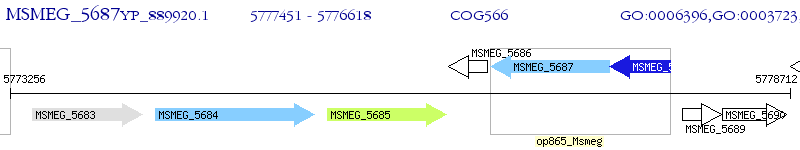
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5687 | - | - | 100% (277) | 23s ribosomal RNA methyltransferase |
| M. smegmatis MC2 155 | MSMEG_3790 | - | 1e-16 | 30.42% (240) | RNA methyltransferase, TrmH family protein |
| M. smegmatis MC2 155 | MSMEG_6073 | - | 3e-14 | 31.67% (180) | RNA methyltransferase, TrmH family protein, group 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0905 | - | 1e-110 | 73.36% (274) | rRNA methyltransferase |
| M. gilvum PYR-GCK | Mflv_1715 | - | 1e-118 | 79.93% (269) | tRNA/rRNA methyltransferase (SpoU) |
| M. tuberculosis H37Rv | Rv0881 | - | 1e-110 | 73.36% (274) | rRNA methyltransferase |
| M. leprae Br4923 | MLBr_00324 | - | 4e-16 | 29.28% (181) | putative methyltransferase |
| M. abscessus ATCC 19977 | MAB_0881 | - | 1e-111 | 74.54% (271) | putative tRNA/rRNA methyltransferase |
| M. marinum M | MMAR_4651 | - | 1e-106 | 72.00% (275) | rRNA methyltransferase |
| M. avium 104 | MAV_1009 | - | 1e-112 | 74.91% (271) | SpoU rRNA methylase family protein |
| M. thermoresistible (build 8) | TH_1372 | - | 1e-117 | 79.34% (271) | POSSIBLE RRNA METHYLTRANSFERASE (RRNA METHYLASE) |
| M. ulcerans Agy99 | MUL_0269 | - | 1e-104 | 71.85% (270) | rRNA methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5036 | - | 1e-120 | 80.51% (272) | tRNA/rRNA methyltransferase (SpoU) |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1715|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5036|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1372|M.thermoresistible__bu --------------------------------------------------
MSMEG_5687|M.smegmatis_MC2_155 --------------------------------------------------
Mb0905|M.bovis_AF2122/97 --------------------------------------------------
Rv0881|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4651|M.marinum_M --------------------------------------------------
MUL_0269|M.ulcerans_Agy99 --------------------------------------------------
MAV_1009|M.avium_104 MPDSDARLASDLSLAVMRLARQLRFRNPSSPVSLSQLSALAMLANEGPMT
MAB_0881|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00324|M.leprae_Br4923 --------------------------------------------------
Mflv_1715|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5036|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1372|M.thermoresistible__bu --------------------------------------------------
MSMEG_5687|M.smegmatis_MC2_155 --------------------------------------------------
Mb0905|M.bovis_AF2122/97 --------------------------------------------------
Rv0881|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4651|M.marinum_M --------------------------------------------------
MUL_0269|M.ulcerans_Agy99 --------------------------------------------------
MAV_1009|M.avium_104 PGALAVRERVRPPSMTRVIASLAEMGLVDRTPHPVDGRQVLVSVSEAGAE
MAB_0881|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00324|M.leprae_Br4923 --------------------------------------------------
Mflv_1715|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5036|M.vanbaalenii_PYR-1 -------------------------------------------MIANNVE
TH_1372|M.thermoresistible__bu ----------------------------VHTLSGPNPDDPDGPDVTDGPD
MSMEG_5687|M.smegmatis_MC2_155 ---------------------------------------------MSPLQ
Mb0905|M.bovis_AF2122/97 ------------------------------------MTEGRCAQHPDGLD
Rv0881|M.tuberculosis_H37Rv ------------------------------------MTEGRCAQHPDGLD
MMAR_4651|M.marinum_M ------------------------------------MTA---HSHVRDVD
MUL_0269|M.ulcerans_Agy99 --------------------------------------------------
MAV_1009|M.avium_104 LVKANRRARQEWLAKRLVHAGRRRARNPAPRGRSDAGPGRRRPVNTDALD
MAB_0881|M.abscessus_ATCC_1997 ------------------------------------------------ML
MLBr_00324|M.leprae_Br4923 -------------------------------------------MEGRGPT
Mflv_1715|M.gilvum_PYR-GCK MVDITDPADPRLDPFRDLNSVDRRPDLPSGKGLVIAEG-VLVVQRMLASR
Mvan_5036|M.vanbaalenii_PYR-1 VVDVTDPADPRLDDFRDLNSVDRRPDLPSGKGLVIAEG-VLVVQRMLASR
TH_1372|M.thermoresistible__bu VTDITDPADPRVDDFRDLNSVDRRPDLPGGKGLVIAEG-VLVVQRMLASR
MSMEG_5687|M.smegmatis_MC2_155 VIDIDDPADPRIDDFRDLNSVDRRPDLPTGKALVIAEG-VLVVQRMLASR
Mb0905|M.bovis_AF2122/97 VQDVCDPDDPRLDDFRDLNSIDRRPDLPTGKALVIAEG-VLVVQRMLASR
Rv0881|M.tuberculosis_H37Rv VQDVCDPDDPRLDDFRDLNSIDRRPDLPTGKALVIAEG-VLVVQRMLASR
MMAR_4651|M.marinum_M VRDVSDPDDPCLDDFRDLNSIDRRPDLPSGKGLVIAEG-VLVVQRMLASR
MUL_0269|M.ulcerans_Agy99 MRDVSDPDDPCLDDFRDLNSIDRRPDLPSGKGLVIAEG-VLVVQRMLASR
MAV_1009|M.avium_104 VRDVDDPDDPRLDDFRDLNSVDRRPDLPSGKGLVIAEG-VLVVQRMLASR
MAB_0881|M.abscessus_ATCC_1997 VIDVHDPADSRLDSFRDLNSVDRRPDLPSGKGLVIAEG-VLVVQRMLASR
MLBr_00324|M.leprae_Br4923 PPAYLRPNHPAAK--RNQSSPHRPVKRTDETETVLGRNPVLECLRAGVPA
* .. . *: .* .* . . . *:... ** * ..
Mflv_1715|M.gilvum_PYR-GCK FVPRALLG--TDRRLTELGTDLDG----LAAPFYRATAEVMAEAIGFHLN
Mvan_5036|M.vanbaalenii_PYR-1 FVPRALLG--TDRRLTELGTDLAG----LDAPFYRASAEVMADVVGFHLN
TH_1372|M.thermoresistible__bu FTPRAFLG--TDRRLTELRADLAA----VDAPFYRADAEVMAEVVGFHLN
MSMEG_5687|M.smegmatis_MC2_155 FIPRALLG--TDRRLGELAADLDRYPQYADVPFYRASAEVMAEAIGFHLN
Mb0905|M.bovis_AF2122/97 FTPLALFG--TDRRLAELKDDLAG----VGAPYYRASADVMARVIGFHLN
Rv0881|M.tuberculosis_H37Rv FTPLALFG--TDRRLAELKDDLAG----VGAPYYRASADVMARVIGFHLN
MMAR_4651|M.marinum_M FSPRALLG--TDRRLLELKDDLAG----VGVPYYRVSPDVMAQVVGFHLN
MUL_0269|M.ulcerans_Agy99 FSPRALLG--TDRRLLELKEDLAG----VGAPYYRVSPDVMAQVVGFHLN
MAV_1009|M.avium_104 FRPHALLG--TERRLAELRDDLAG----VPVPYYRTSADVMARVVGFHLN
MAB_0881|M.abscessus_ATCC_1997 FRPLALLG--TDRRLSELAADLEP----VDVPYYRASAEVMAEVVGFHLN
MLBr_00324|M.leprae_Br4923 TALYVALGTEVDKRLTESVMRAAD----VGVAILEVPRTYLDRITNNHLH
. :* .::** * .. .. : . **:
Mflv_1715|M.gilvum_PYR-GCK RGVLASASRAPELTVAEVLDGAR-----TVAVLEGVNDHENLGSVFRNAA
Mvan_5036|M.vanbaalenii_PYR-1 RGVLASASRAPELTVPHVLDGAR-----TVAVLEGVNDHENLGSVFRNAA
TH_1372|M.thermoresistible__bu RGVLASAARPTELTVPQVLDGAR-----TVAVLEGVNDHENLGSIFRNAA
MSMEG_5687|M.smegmatis_MC2_155 RGVLASAPRAAELTVDDVIADAR-----TIAVLEGVNDHENLGSIFRNAA
Mb0905|M.bovis_AF2122/97 RGVLAAARRVPEPSVAQVVAGAR-----TVAVLEGVNDHENLGSIFRNAA
Rv0881|M.tuberculosis_H37Rv RGVLAAAGRVPEPSVAQVVAGAR-----TVAVLEGVNDHENLGSIFRNAA
MMAR_4651|M.marinum_M RGVLAAASRVVEPSVGQVVDGAR-----TIAVLEGVNDHENLGSIFRNAA
MUL_0269|M.ulcerans_Agy99 RGVLAAASRVVEPSVGQVVDGAR-----TIAVLEGVNDHENLSSIFRNAA
MAV_1009|M.avium_104 RGVLAAARRVPEPGVAELLAGSR-----TVAVLEGVNDHENLGAIFRNAA
MAB_0881|M.abscessus_ATCC_1997 RGVLAASRRPEELPAADLLQTAR-----TVAVLEGVNDHENLGSIFRNAA
MLBr_00324|M.leprae_Br4923 QGIALQVPPYHYVHSDDLLAAATDSPPALLVALDNISDPRNLGAIVRSVA
:*: .:: : :..*:.:.* .**.::.*..*
Mflv_1715|M.gilvum_PYR-GCK GLGVDAVIFGSGCADPLYRRAVRVSMGHALLVPFARAANWPADLNILRDN
Mvan_5036|M.vanbaalenii_PYR-1 GLGVDGVIFGSGCADPLYRRAVRVSMGHALLVPFAKAANWPGDLELLRDQ
TH_1372|M.thermoresistible__bu GLGVDAVIFGSGCADPLYRRAVRVSMGHALLVPYARARHWPADLRLLRDN
MSMEG_5687|M.smegmatis_MC2_155 GLAVDAVIFGAGCADPLYRRAVRVSMGHALLVPFARASSWPTDLELLRDK
Mb0905|M.bovis_AF2122/97 GLSVDAVVFGTGCADPLYRRAVRVSMGHALLVPYARAADWPTELMTLKES
Rv0881|M.tuberculosis_H37Rv GLSVDAVVFGTGCADPLYRRAVRVSMGHALLVPYARAADWPTELMTLKES
MMAR_4651|M.marinum_M GLGVDAVIFGSGCADPLYRRAVRVSMGHALLVPYARATDWPAELVALKER
MUL_0269|M.ulcerans_Agy99 GLGVHAVIFGSGCADPLYRRAVRVSMGHALLVPYARATDWPAELVALKER
MAV_1009|M.avium_104 GLGVDAVIFGSGCADPLYRRAVRVSMGHALLVPYARATDWPAELKTLKEK
MAB_0881|M.abscessus_ATCC_1997 GMGVDAVLFGSGCADPLYRRAVRVSMGHALLVPFARLPEWPKSLYRLRDS
MLBr_00324|M.leprae_Br4923 AFSGHGILIPQRRSASVTAVAWRTSAGAAARIPVARATNLTRALKVWADQ
.:. ..::: : .: * *.* * * :* *: . * :
Mflv_1715|M.gilvum_PYR-GCK DFRLLAMTPDPTVPTLADAVSGLT-EERIAILVGAEGPGLKEHTMRSADV
Mvan_5036|M.vanbaalenii_PYR-1 GFRLLAMTPDPTVQTLAQAMSGLA-EQRVAILVGAEGPGLKEHTMRASDV
TH_1372|M.thermoresistible__bu GFQLLAMTPNPKAETLAAVMPRLA-AQKVAVLVGAEGPGLAESTMRAADV
MSMEG_5687|M.smegmatis_MC2_155 GFRLLAMTPDPSAATLSDAMVQLE-GDRVAILVGAEGPGLTERTMRASDV
Mb0905|M.bovis_AF2122/97 GFRLLAMTPHGNACKLPEAIAAVS-HERIALLVGAEGPGLTAAALRISDV
Rv0881|M.tuberculosis_H37Rv GFRLLAMTPHGNACKLPEAIAAVS-HERIALLVGAEGPGLTAAALRISDV
MMAR_4651|M.marinum_M GFRLLALTPKGDARALPEAMDAVG-DQPVALLVGAEGPGLTAAALRISDL
MUL_0269|M.ulcerans_Agy99 GFRLLALTPKGDARALPEAMDAVG-DQPVALLVGAEGPGLTAAALRISDL
MAV_1009|M.avium_104 GFRLLAMTPQGSACGLAEAMTAAR-GDRVAVLVGAEGPGLTPAALRLSDV
MAB_0881|M.abscessus_ATCC_1997 GFRLLAMTPAGDSKALADAIPPLR-AEKVALMVGAEGPGLTDVALRASDL
MLBr_00324|M.leprae_Br4923 GLRVIGLDHDGDT-----ALDDLDGTDPLAVVVGSEGKGLSRLVRHSCDE
.::::.: .: : :*::**:** ** . : .*
Mflv_1715|M.gilvum_PYR-GCK RVRIPMARGTDSLNVATAAALAFYERDR--SHR------------
Mvan_5036|M.vanbaalenii_PYR-1 RVRIPMSRGTDSLNVATAAALAFYERDR--LGR------------
TH_1372|M.thermoresistible__bu RVRIPMSRGTDSLNVATAAALAFYERSR--PSG------------
MSMEG_5687|M.smegmatis_MC2_155 RVRIPMSRGTDSLNVATAAALAFYERAR--LSH------------
Mb0905|M.bovis_AF2122/97 RVRIPMSRGTDSLNVATAAALAFYERTR--SGHHIGPGT------
Rv0881|M.tuberculosis_H37Rv RVRIPMSRGTDSLNVATAAALAFYERTR--SGHHIGPGT------
MMAR_4651|M.marinum_M RVRIPMSRGTDSLNVATAAALAFYERAR--LGQ------------
MUL_0269|M.ulcerans_Agy99 RVRIPMSRGTDSLNVATAAALAFYERAR--LGQ------------
MAV_1009|M.avium_104 RVRIPMSRGTDSLNVATAAALAFYERSRPVGGHRAAAGGGDGQSG
MAB_0881|M.abscessus_ATCC_1997 RVRIPMANGTDSLNVATAAALAFYERAR--LAP------------
MLBr_00324|M.leprae_Br4923 VVSIPMAGQVESLNASVAVGVVLAEIAR--QRRL-----------
* ***: .:***.:.*..:.: * *