For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAVQLVTSDLVGVALRSVDGLEVSLPQFRLLRVLDEVGEAGATRCAEMLGIGGSSITRLVDRLHASGHLV RRPDPANRSAVVLVLTDEGRRLVRRVESRRRRELGKALDRLSPKERSQCIAALERLHEVLQAPEDPRIPY *
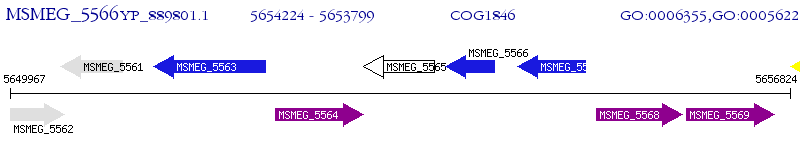
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5566 | - | - | 100% (141) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6814 | - | 2e-10 | 29.82% (114) | helix-turn-helix, Fis-type |
| M. smegmatis MC2 155 | MSMEG_4471 | - | 2e-09 | 27.73% (119) | MarR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1439 | - | 2e-14 | 31.50% (127) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_0628 | - | 2e-07 | 25.00% (120) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1404 | - | 2e-14 | 31.50% (127) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00550 | - | 1e-15 | 35.34% (116) | putative marR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1021 | - | 6e-14 | 34.19% (117) | MarR family transcriptional regulator |
| M. marinum M | MMAR_2213 | - | 2e-17 | 37.07% (116) | transcriptional regulatory protein |
| M. avium 104 | MAV_3374 | - | 2e-14 | 34.48% (116) | transcriptional regulator, MarR family protein |
| M. thermoresistible (build 8) | TH_3245 | - | 3e-08 | 26.26% (99) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1800 | - | 1e-17 | 37.07% (116) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4344 | - | 6e-08 | 33.93% (112) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1439|M.bovis_AF2122/97 --------------------------------------------------
Rv1404|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 MARTAIANKAISEYLRPRGSLGSGTEANTSRNSDPMSSLRSPALQWSNTS
MLBr_00550|M.leprae_Br4923 --------------------------------------------------
MAV_3374|M.avium_104 --------------------------------------------------
MAB_1021|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5566|M.smegmatis_MC2_155 --------------------------------------------------
TH_3245|M.thermoresistible__bu --------------------------------------------------
Mflv_0628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4344|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1439|M.bovis_AF2122/97 --------------------------------------------------
Rv1404|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 GTDDTGNTETVGTGNSRPTTNSNQLDRSPDSPCLHCGPNSAAPKIINPAH
MLBr_00550|M.leprae_Br4923 --------------------------------------------------
MAV_3374|M.avium_104 --------------------------------------------------
MAB_1021|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_5566|M.smegmatis_MC2_155 --------------------------------------------------
TH_3245|M.thermoresistible__bu --------------------------------------------------
Mflv_0628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4344|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1439|M.bovis_AF2122/97 ---------------MMPTEYPATAEESVDVITDALLTASRLLVAISAHS
Rv1404|M.tuberculosis_H37Rv ---------------MMPTEYPATAEESVDVITDALLTASRLLVAISAHS
MMAR_2213|M.marinum_M ----------------MSTENATAAEESLDMITDALLTASRLLVGISAHS
MUL_1800|M.ulcerans_Agy99 KPANQQRRSPGGRGPLMSTENATAAEESLDMITDALPTASRLLVGISAHS
MLBr_00550|M.leprae_Br4923 ----------------MVTDDATVADESLDDITDALLTASRLLVAVSAHS
MAV_3374|M.avium_104 ----------------MAADH-TTVDESLDAITDALLTASRLLMAISARS
MAB_1021|M.abscessus_ATCC_1997 ----------------MTVDD-VDLDESLDALADSLLVASRLLLAISARS
MSMEG_5566|M.smegmatis_MC2_155 --------------------------------MVAVQLVTSDLVGVALRS
TH_3245|M.thermoresistible__bu -----------------MEERSSGIAALHQRIPFLLSQLGAHLAEDFVRR
Mflv_0628|M.gilvum_PYR-GCK --MTTEKLTMAQQVAAKTREAGPDHDPSSLALPLALYRAVTAFGRVAVDE
Mvan_4344|M.vanbaalenii_PYR-1 -------------------------MDAQPGTAVLMYIAHREVESRVMAA
: .
Mb1439|M.bovis_AF2122/97 IAQVDE-NITIPQFRTLVILS-NHGPINLATLATLLGVQPSATGRMVDRL
Rv1404|M.tuberculosis_H37Rv IAQVDE-NITIPQFRTLVILS-NHGPINLATLATLLGVQPSATGRMVDRL
MMAR_2213|M.marinum_M IAQVDE-NITIPQFRTLVILS-NRGPVNLATLANLLGVQPSATGRMVDRL
MUL_1800|M.ulcerans_Agy99 IAQVDE-NITIPQFRTLVILS-NRGPVNLATLANLLGVQPSATGRMVDRL
MLBr_00550|M.leprae_Br4923 IGQVDE-TITFPQFRTLVILS-NRGPVNLATLAGLLGVQPSATGRMVDRL
MAV_3374|M.avium_104 VGQVDE-TITIPQFRTLVILS-NRGPVNLATLAGLLGVQPSATGRMVDRL
MAB_1021|M.abscessus_ATCC_1997 IAQVDE-TITFPQFRTLVILF-NEGPINLATLAQSLGVQPSTTGRMVDRL
MSMEG_5566|M.smegmatis_MC2_155 VDGL---EVSLPQFRLLRVLD-EVGEAGATRCAEMLGIGGSSITRLVDRL
TH_3245|M.thermoresistible__bu LEPFG---IEPRIYAVLTALS-EVDGQSQRELSAQLGIHRNAMVAVVDKM
Mflv_0628|M.gilvum_PYR-GCK LTPVD---LSMSQFNVLTVLKRADGPITMGALADSISVRQASLTSVVDTL
Mvan_4344|M.vanbaalenii_PYR-1 LARSGVGDVTVAQSRLLQRLD--PGGMRLTDLAEQARVTKQTAGALVDQL
: : * * . : : : :** :
Mb1439|M.bovis_AF2122/97 VGAELIDRLPHPTSRRELLAALTKRGRDVVRQVTEHRRTEIARIVEQMAP
Rv1404|M.tuberculosis_H37Rv VGAELIDRLPHPTSRRELLAALTKRGRDVVRQVTEHRRTEIARIVEQMAP
MMAR_2213|M.marinum_M VSAGMIDRQPHPTSRRELLAVLTKRGRDLVRRVTAHRRDEIARIVEQMPP
MUL_1800|M.ulcerans_Agy99 VSAGMIDRQPHPTSRRELLAVLTKRGRDLVRRVTAHRRDEIARIVEQMPP
MLBr_00550|M.leprae_Br4923 VAARLIDRLPHPTSRRELLVALTKRGREVVRRVTAQRRVEIARIVEKMPP
MAV_3374|M.avium_104 VAAGLIDRLPHPTSRRELLAALTTRGRDVVHQVTAHRRAEIAAIVAKMPP
MAB_1021|M.abscessus_ATCC_1997 VSAGLIDRRPHPRSRRELVAELTAHGEGVVREVMAQRRAEIAEIAARMPA
MSMEG_5566|M.smegmatis_MC2_155 HASGHLVRRPDPANRSAVVLVLTDEGRRLVRRVESRRRRELGKALDRLSP
TH_3245|M.thermoresistible__bu ESLGLVKRLPHPADRRAFAVTLTDKARELLPALDAEGRAQEDAVTAAMTP
Mflv_0628|M.gilvum_PYR-GCK TKAGFVTRKVNPRDRRSVVVAISKKGDQFMTEFLPGHYDFLQRLFAGINP
Mvan_4344|M.vanbaalenii_PYR-1 ERAGYVTRQPDPSDARARLVTLTDKGAEMCRRAGAEVARVEQEWRQHLGD
: * .* . :: .. . :
Mb1439|M.bovis_AF2122/97 AERHGLVRALTAFTEAGGEPDARYEIE---
Rv1404|M.tuberculosis_H37Rv AERHGLVRALTAFTEAGGEPDARYEIE---
MMAR_2213|M.marinum_M AERQGLVRALTAFTAAGGEPDANVEMDL--
MUL_1800|M.ulcerans_Agy99 AERQGLVRALTAFTAAGGEPDANVEMDL--
MLBr_00550|M.leprae_Br4923 SERLGLVRALTAFSTAGGEPDVRLDIISI-
MAV_3374|M.avium_104 AERHGLVRALTAFTAAGGEPDVHLDAEIDL
MAB_1021|M.abscessus_ATCC_1997 RERRGLVRALDAFTAAGGESASPVDV----
MSMEG_5566|M.smegmatis_MC2_155 KERSQCIAALERLHEVLQAPEDPRIPY---
TH_3245|M.thermoresistible__bu AERRETLRLLQKMSSTVGLVPGVHPKLG--
Mflv_0628|M.gilvum_PYR-GCK RHRQQLLARLNELIDCLENYPSGQRTS---
Mvan_4344|M.vanbaalenii_PYR-1 DAYGALRAALLSLREITDPYL---------
* :