For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSLRPAEQNSAGAGTVLKPWHDTTVPAAERVELLLAELSLEEKVAQLGSRWVSNDRGAREDDRDAHDFAE TAEQEASLNVAPMEDAFALSGSVPLHEAARHGLGHLTRVYGSRPVTPAEGAAELMRQQRTVLEQSRLDIP ALVHEECLTGFTAYGATVYPAAIAWGATFDPPLVREMAAAIGRDMAALGVHQGLSPVLDVVRDYRWGRVE ETMGEDPYVVSTLGAAYIEGLQSAGVIATLKHFAGYSASRAGRNHAPVPMGRRELADAIFPPFEVAVTVA GAGSVMSAYNDIDGVPASADAWLLTDVLRGQWGFRGTVVSDYWSVPFLAMMHRVAADADESGAAALAAGV DVELPDTVGFGTNIVERVRNGQLPVEFVDRAVRRLLLQKVQLGLLDPGWTPEASVAGAAETDLDAPANRD IARRLAERSVILLDAGSALPLDGPGTQAPGRIAVVGPCAADARTFMGCYAFPNHVLPRYPGRGLGVAVPT VVDALADELSGAQIRYEQGCGVQDPDRSGFTAAVDAAKSADLCIAVVGDLAGMFGNGSSGEGSDAEDLRL PGVQADLLEELLATGVPVVVVVVSGRPYALGAVAGRAAGLIQAFMPGEEGGAAIAGVLSGRVQPGGKLPV QIPRTPGGQPGTYLQPPLGTLGADVSSVDPTPLYPFGYGRSYTTFEVDDLRISSERVPTDGDWTVSVRVR NTGPRDGDEVVQLYLEDPVASVARPTRQLMGYARVTVPAASAVTVTFGLHADRTAFTGRDLTRIVEAGQV DLLVGTSAADIACRGTVRLTGPTRTVGSDRTMLTPVTVVPEDPR*
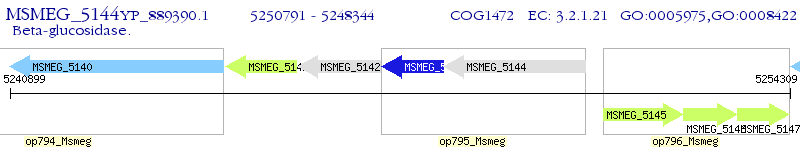
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5144 | - | - | 100% (815) | xylosidase/arabinosidase |
| M. smegmatis MC2 155 | MSMEG_2556 | - | 3e-70 | 33.23% (662) | beta-glucosidase |
| M. smegmatis MC2 155 | MSMEG_0361 | - | 1e-07 | 30.09% (226) | glycosyl hydrolase family protein 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0192 | bglS | 5e-65 | 30.76% (699) | beta-glucosidase BGLS |
| M. gilvum PYR-GCK | Mflv_0415 | - | 2e-12 | 29.80% (245) | Beta-N-acetylhexosaminidase |
| M. tuberculosis H37Rv | Rv0186 | bglS | 5e-65 | 30.76% (699) | beta-glucosidase |
| M. leprae Br4923 | MLBr_02569 | - | 2e-09 | 30.68% (251) | putative secreted glycosyl hydrolase |
| M. abscessus ATCC 19977 | MAB_4458c | - | 6e-09 | 27.46% (244) | putative secreted hydrolase |
| M. marinum M | MMAR_0430 | bglS | 2e-64 | 30.62% (699) | beta-glucosidase BglS |
| M. avium 104 | MAV_4994 | - | 2e-63 | 31.25% (688) | glycosyl hydrolase family protein 3 |
| M. thermoresistible (build 8) | TH_1910 | lpqI | 6e-10 | 25.12% (410) | PROBABLE CONSERVED LIPOPROTEIN LPQI |
| M. ulcerans Agy99 | MUL_1080 | bglS | 6e-63 | 30.22% (695) | beta-glucosidase BglS |
| M. vanbaalenii PYR-1 | Mvan_0256 | - | 1e-12 | 30.74% (257) | Beta-N-acetylhexosaminidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1910|M.thermoresistible__bu --------------------------------------------------
MLBr_02569|M.leprae_Br4923 --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 --------------------------------------MTDDERFSLLVG
Rv0186|M.tuberculosis_H37Rv --------------------------------------MTDDERFSLLVG
MMAR_0430|M.marinum_M ------------------MSSSGHNAPADVRARQVESEMTDEERFSLLLG
MUL_1080|M.ulcerans_Agy99 --------------------------------------MTDEERFSLLLG
MAV_4994|M.avium_104 ------------------------MKDPDERARTVEAQMTDDERFSLLVA
MSMEG_5144|M.smegmatis_MC2_155 MTSLRPAEQNSAGAGTVLKPWHDTTVPAAERVELLLAELSLEEKVAQLGS
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 --------MANPRTAPEALVGVLAASALLVGCTSSTAPEPTPHVLHATPS
TH_1910|M.thermoresistible__bu -----------------------LTLACSPAAERPAEPEPDPPAVVATP-
MLBr_02569|M.leprae_Br4923 ------------MAFPRTLIVLAAASALVVTCGHDVAQAPDS--------
MAB_4458c|M.abscessus_ATCC_199 --------------MRAGTVLTGLVVAALTACGGNASE------------
Mb0192|M.bovis_AF2122/97 LTGASDLWPVRDERIPQGVPMCAGYVPGIPRLGVPALLMSDAGLGVTNPG
Rv0186|M.tuberculosis_H37Rv LTGASDLWPVRDERIPQGVPMCAGYVPGIPRLGVPALLMSDAGLGVTNPG
MMAR_0430|M.marinum_M VMGAGDLWPLRDERIPADVPMSAGYVPGIARLGVPALLMSDAGLGVTNPG
MUL_1080|M.ulcerans_Agy99 VMGAGDLWPLRDERIPADVPMSAGYVPGIARLGVPALLMTDAGLGVTNPG
MAV_4994|M.avium_104 VMGAGDMWPVRDERIPADVPMSAGYVPGVPRLGVPPLLMSDAGLGVTNPG
MSMEG_5144|M.smegmatis_MC2_155 RWVSNDRGAREDDRDAHDFAETAEQEASLNVAPMEDAFALSGSVPLHEAA
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 M-------------------------------------------------
TH_1910|M.thermoresistible__bu --------------------------------------------------
MLBr_02569|M.leprae_Br4923 --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 YR------------------------------------------------
Rv0186|M.tuberculosis_H37Rv YR------------------------------------------------
MMAR_0430|M.marinum_M YR------------------------------------------------
MUL_1080|M.ulcerans_Agy99 YR------------------------------------------------
MAV_4994|M.avium_104 YR------------------------------------------------
MSMEG_5144|M.smegmatis_MC2_155 RHGLGHLTRVYGSRPVTPAEGAAELMRQQRTVLEQSRLDIPALVHEECLT
Mflv_0415|M.gilvum_PYR-GCK ----------------------------------MSTRDKLAQLLMVGVT
Mvan_0256|M.vanbaalenii_PYR-1 ------AAGPALNLPAAPAPAAPACGDPAAIVASMSTRDKLAQRLMVGVT
TH_1910|M.thermoresistible__bu -------AATPLSTPLPAAPPAG-CGDAQSVLASMELRDKLAQLLMVGVN
MLBr_02569|M.leprae_Br4923 -------ASIVAGELTAAALTPACVDLPASVPGTLSTRDKLAQLLMVGVR
MAB_4458c|M.abscessus_ATCC_199 ---------PSKENTSSTKAQAPATAAPAADLACLSTRDRLAQLLMVGIK
Mb0192|M.bovis_AF2122/97 --PGDTATALPAGLALAASFNPVLARSSGKAIGREARSRGFNVQLAGAIN
Rv0186|M.tuberculosis_H37Rv --PGDTATALPAGLALAASFNPVLARSSGKAIGREARSRGFNVQLAGAIN
MMAR_0430|M.marinum_M --PGDTATALPAGLALAASFNPALARSAGEVIGREARSRGFNVQLAGAMN
MUL_1080|M.ulcerans_Agy99 --PGDTATALPAGLALAASFNPALARSAGEVIGREARSRGFNVQLAGAMN
MAV_4994|M.avium_104 --PGDTATALPAGLALAATFDPALARAAGELIGREARSRGFNVQLAGAMN
MSMEG_5144|M.smegmatis_MC2_155 GFTAYGATVYPAAIAWGATFDPPLVREMAAAIGRDMAALGVHQGLSPVLD
. * :
Mflv_0415|M.gilvum_PYR-GCK GAADAR-----------TAAAEYHVGGIMVGSWTDLSMLTDGSLP-DIAN
Mvan_0256|M.vanbaalenii_PYR-1 GAADAR-----------AVVDAHHVGGIMVGSWTDLSMLSDNSLA-DIAA
TH_1910|M.thermoresistible__bu GAADAR-----------AVVNDHRVGGIFVGSWSDLSMLSDGSVA-DIAA
MLBr_02569|M.leprae_Br4923 DAADAK-----------AVVTDHHVGGIMIGSWTDLSMLINGALT-DIAR
MAB_4458c|M.abscessus_ATCC_199 DADDAR-----------ALVRDHHVGGIFVGSWTKKEMLTDGSLH-DVIT
Mb0192|M.bovis_AF2122/97 LARDPRNGRNFEYLSEDPLLSATMAAESIIGIQQQGVIATTKHFS-LNCN
Rv0186|M.tuberculosis_H37Rv LARDPRNGRNFEYLSEDPLLSATMAAESIIGIQQQGVIATTKHFS-LNCN
MMAR_0430|M.marinum_M LARDPRNGRNFEYLSEDPFLTAAMVAESIIGIQQQGVISTVKHFS-LNCN
MUL_1080|M.ulcerans_Agy99 LARDPRNGRNFEYLSEDPFLTAAMVAESIIGIQQQGVISTVKHFS-LNCN
MAV_4994|M.avium_104 LARDPRNGRNFEYLSEDPLLTAAMVAESVAGIQQQGVISTVKHYS-LNCN
MSMEG_5144|M.smegmatis_MC2_155 VVRDYRWGRVEETMGEDPYVVSTLGAAYIEGLQSAGVIATLKHFAGYSAS
. * : . . . * :
Mflv_0415|M.gilvum_PYR-GCK -AGLLPLAVSVDEEGGRVSRLSGLIGPQPSARVLAQTKTPDEVRAIARD-
Mvan_0256|M.vanbaalenii_PYR-1 TAAPLPPAVSVDEEGGRVSRLAGLIGAQPSPRRLAATSSPEQVYQIALE-
TH_1910|M.thermoresistible__bu SAGPFPLAVSVDEEGGRVSRLSGLIGPQPSPRSLARTRSPEEVQVIARE-
MLBr_02569|M.leprae_Br4923 NPGPLPLAVSVDEEGGRVSRLKSLIGMAPSARVLAQTSTVDQVYDMALD-
MAB_4458c|M.abscessus_ATCC_199 -AGPIPAAVSIDEEGGRVSRLKDLIGPAPSARELAATQTPEQVQALSQE-
Mb0192|M.bovis_AF2122/97 ETNRHWLDAVIDPDAHRESDLLAFEIVIERSQPGAVMAAYNKVNGDYAAG
Rv0186|M.tuberculosis_H37Rv ETNRHWLDAVIDPDAHRESDLLAFEIVIERSQPGAVMAAYNKVNGDYAAG
MMAR_0430|M.marinum_M ETNRHWLDAVIDPDAHRESDLLAFEIAIERAQPGAVMAAYNKVNGVYAAA
MUL_1080|M.ulcerans_Agy99 ETNRHWLDAVIDPDAHRESDLLAFEIAIERAQPGAVVAAYNKVNGVYATA
MAV_4994|M.avium_104 ETNRHFLDAVIDPDAHRESDLLAFELAIERAQPGAVMTAYNKVNGVYAAA
MSMEG_5144|M.smegmatis_MC2_155 RAGRNHAPVPMGRRELADAIFPPFEVAVTVAGAGSVMSAYNDIDGVPASA
. . :. : : : . : : :.:
Mflv_0415|M.gilvum_PYR-GCK ---------------RGAKMRNLGITVDFAPVVDVSDAPDN-------TV
Mvan_0256|M.vanbaalenii_PYR-1 ---------------RGRKMRGLGITIDFAPVVDVTDAPDG-------TV
TH_1910|M.thermoresistible__bu ---------------RGQAMRGRGITIDFAPVVDVTDAADN-------SV
MLBr_02569|M.leprae_Br4923 ---------------RGHKMRDLGITIDFAPVVDVTDAPDD-------SV
MAB_4458c|M.abscessus_ATCC_199 ---------------RAQKMKDLGITIDFAPDADVTDGDPD-------GA
Mb0192|M.bovis_AF2122/97 NDHLLNDVLKGAWGYRGWVMSDWGGTPSWECALAGLDQECG-------AQ
Rv0186|M.tuberculosis_H37Rv NDHLLNDVLKGAWGYRGWVMSDWGGTPSWECALAGLDQECG-------AQ
MMAR_0430|M.marinum_M NGELINEVLKGAWGYRGWVMSDWGGTPGWECALGGLDQECG-------AQ
MUL_1080|M.ulcerans_Agy99 NGELINEVLKGAWGYRGWVMSDWGGTPGWECALGGLDQECG-------AQ
MAV_4994|M.avium_104 NSVLLNDVLKDAWAYRGWVMSDWGATPGWECALGGLDQECG-------AQ
MSMEG_5144|M.smegmatis_MC2_155 DAWLLTDVLRGQWGFRGTVVSDYWSVPFLAMMHRVAADADESGAAALAAG
*. : . .
Mflv_0415|M.gilvum_PYR-GCK IGDRSFG----ADPTTVTDYAGAYAQGLRDAGVLPVLKHFPGHGSG----
Mvan_0256|M.vanbaalenii_PYR-1 IGDRSFG----ADPTAVVDYAGAYARGLRDAGVLPVLKHFPGHGSG----
TH_1910|M.thermoresistible__bu IGDRSFG----ADPETVTAYAGAYARGLREGGVMPVLKHFPGHGRA----
MLBr_02569|M.leprae_Br4923 IGGRSFS----SDPAVVTAYAGAYARGLRDAGVLPVLKHFPGHGHG----
MAB_4458c|M.abscessus_ATCC_199 IGDRSFS----GDPQKVAEYAEASVRGYQAGGVRAVVKHFPGHGHA----
Mb0192|M.bovis_AF2122/97 IDAVLWQSEAFTDRLRAAYADGNLPKGRLSDMVRRILRSMFAVGIDRWKP
Rv0186|M.tuberculosis_H37Rv IDAVLWQSEAFTDRLRAAYADGNLPKGRLSDMVRRILRSMFAVGIDRWKP
MMAR_0430|M.marinum_M IDAILWQSEAFGDPLRAAYHEGRLPKARLSEMVRRILRSMFAVGVDRWEP
MUL_1080|M.ulcerans_Agy99 IDAILWQSEAFGDPLRAAYHEGRLPKARLSEMVRRILRSMFAVGVDRWEP
MAV_4994|M.avium_104 IDALLWQAESFGAPLRDAYADGRLPKDRLSDMVRRILRSIFAVGVDRSDR
MSMEG_5144|M.smegmatis_MC2_155 VDVELPDTVGFGTNIVERVRNGQLPVEFVDRAVRRLLLQKVQLGLLDPGW
:. * :: *
Mflv_0415|M.gilvum_PYR-GCK -----------------SGDSHVGSVVAPSLEQLQASDLVPYRTLAAQAP
Mvan_0256|M.vanbaalenii_PYR-1 -----------------SGDTHAGSVVTPPLGDLVARDLVPYRTLTTQGP
TH_1910|M.thermoresistible__bu -----------------SGDSHASGVVTPPLADLEATDLVPYRALVGEQ-
MLBr_02569|M.leprae_Br4923 -----------------SGDSHTGGVTTPPLQDLQTNDLVPYRTLVADAP
MAB_4458c|M.abscessus_ATCC_199 -----------------SGDSHTNGVVTPPLEQLKDNDLVPYRSLVTSG-
Mb0192|M.bovis_AF2122/97 APAPDMN----------AHNEIAAQMARQGIVLLQNRGLLPLAPESAGR-
Rv0186|M.tuberculosis_H37Rv APAPDMN----------AHNEIAAQMARQGIVLLQNRGLLPLAPESAGR-
MMAR_0430|M.marinum_M APAPDMD----------AHDEVAAEIARQGIVLLQNRGVLPLTPAPGTR-
MUL_1080|M.ulcerans_Agy99 APAPDMD----------AHDEVAAEIARQGIVLLQNRGVLPLRPAPGTR-
MAV_4994|M.avium_104 APAPDLN----------AHNDIALRIARQGIVLLTNRGLLPLRPDSPAR-
MSMEG_5144|M.smegmatis_MC2_155 TPEASVAGAAETDLDAPANRDIARRLAERSVILLDAGSALPLDGPGTQAP
: . :. : * . :*
Mflv_0415|M.gilvum_PYR-GCK VGVMVGHVEVPGLTGSDPASLSP-QAYGLLRSGG-YGGPPFGGVVFTDDL
Mvan_0256|M.vanbaalenii_PYR-1 VGVMVGHMEVPGLTGSEPASLSP-PAYALLRSGD-YGGPPFGGPVFTDDL
TH_1910|M.thermoresistible__bu VAVMVGHLQVPGLTGGEPASLSP-DAYRLLRSMG-YHGP-----VFTDDL
MLBr_02569|M.leprae_Br4923 VGVMLGHLDVPGLTGDDPASLSP-AAVQLLRTGANYGAPPFNGPVFSDDV
MAB_4458c|M.abscessus_ATCC_199 AAVMIGHMQVPGLTGDDPASLSG-AAVDLLRKGTGYGAPAYDGVIFTDDL
Mb0192|M.bovis_AF2122/97 IAVIGGYAHLGVPAGYGSSAVTPPGGYAGVIPIGGSGLAAGLRNLYLLPS
Rv0186|M.tuberculosis_H37Rv IAVIGGYAHLGVPAGYGSSAVTPPGGYAGVIPIGGSGLAAGLRNLYLLPS
MMAR_0430|M.marinum_M IAVIGGYAQLGVVAGYGSSAVVPPGGYAEVIPIGGAGLTAGVRNLYLLGS
MUL_1080|M.ulcerans_Agy99 IAVIGGYAQLGVLAGYGSSAVVPPGGYAEVIPIGGAGLTPGVRNLYLLGS
MAV_4994|M.avium_104 IAVIGGYAQLGVPAGFGSSTVVPAGGYAAVVPIGGTALEAGLRNLYLLPS
MSMEG_5144|M.smegmatis_MC2_155 GRIAVVGPCAADARTFMGCYAFPNHVLPRYPGRGLGVAVPTVVDALADEL
: .
Mflv_0415|M.gilvum_PYR-GCK SGMQAISDRLG-----------VAEAVLRALQAGADVALWLS------TG
Mvan_0256|M.vanbaalenii_PYR-1 SGMQAITDRLG-----------VADAVLRSLQAGADVALWLS------TA
TH_1910|M.thermoresistible__bu SGMRAITDRYG-----------VAEAVLRSLQAGADVALWLT------TA
MLBr_02569|M.leprae_Br4923 SSMAAISDRYG-----------LADAVLRTLQAGTDIALWVS------TQ
MAB_4458c|M.abscessus_ATCC_199 SSMAAITDRFP-----------IEEAVLKSIKAGVDWALWVS------TE
Mb0192|M.bovis_AF2122/97 SPLSELRKRLPNAQFEFDPGINPAEAVLAARRADIAIVFAIR------AE
Rv0186|M.tuberculosis_H37Rv SPLSELRKRLPNAQFEFDPGINPAEAVLAARRADIAIVFAIR------AE
MMAR_0430|M.marinum_M SPLDELRTQFGNAELEFDPGINPAEAVAVARRADVAVVFAIR------AE
MUL_1080|M.ulcerans_Agy99 SPLDELRTQFGNAELEFDPGINPAEAVAVARRADVAVVFAIR------AE
MAV_4994|M.avium_104 SPLQELRKLLPENTIDFDPGISPAEAVAAAQRADIAIVFAVR------AE
MSMEG_5144|M.smegmatis_MC2_155 SGAQIRYEQGCGVQDPDRSGFTAAVDAAKSADLCIAVVGDLAGMFGNGSS
* . : . : :
Mflv_0415|M.gilvum_PYR-GCK EVPAVLDRLEKAVGAGELTMSGVDGAVTRIVAMKGPSPRCGG--------
Mvan_0256|M.vanbaalenii_PYR-1 EVPAVLDRLEQAVAAGELTMPRIDEAVTRVIAMKGPSPRCGG--------
TH_1910|M.thermoresistible__bu EVPAVLDRLEAAVAAGELSEARVDESVLRLAGAKGYTPRC----------
MLBr_02569|M.leprae_Br4923 TVPAVLDRLEKVVSAGELAMSAVDASVVRVATIKGPGPGCGR--------
MAB_4458c|M.abscessus_ATCC_199 KVGSVLDRLESAYNAGELNADAINASVRRVLAYKG-VPACG---------
Mb0192|M.bovis_AF2122/97 GEGFDSADLSLPWGQDALIAAVASANANTVVVLETGNPVTMPWRD-SVNA
Rv0186|M.tuberculosis_H37Rv GEGFDSADLSLPWGQDALIAAVASANANTVVVLETGNPVTMPWRD-SVNA
MMAR_0430|M.marinum_M GEGFDSADLTLPWGQDAVISAVAAANPNTVVVLETGNPVAMPWQD-SVNA
MUL_1080|M.ulcerans_Agy99 GEGFDSADLTLPWGQDAVISAVAAANPNTVVVLEGGNPVAMPWQD-SVNA
MAV_4994|M.avium_104 GEGFDLADLSLPSGQDELIGAVAAANPNTVVVLQTGNPVTMPWLG-AVNA
MSMEG_5144|M.smegmatis_MC2_155 GEGSDAEDLRLPGVQADLLEELLATGVPVVVVVVSGRPYALGAVAGRAAG
* : : *
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1910|M.thermoresistible__bu --------------------------------------------------
MLBr_02569|M.leprae_Br4923 --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 IMQAWYPGQAGGQAVAEIVTGQVNPSGRLPITFPVDLGQTPRSQPRELGA
Rv0186|M.tuberculosis_H37Rv IMQAWYPGQAGGQAVAEIVTGQVNPSGRLPITFPVDLGQTPRSQPPELGA
MMAR_0430|M.marinum_M IVQAWYPGQAGARVIAEVLAGRVNPCGRLPITFPVGLGQTPRPELPGYGA
MUL_1080|M.ulcerans_Agy99 IVQAWYPGQAGARVIAEVLAGRVNPCGRLPITFPVGLGQTPRPELPGYGA
MAV_4994|M.avium_104 VVQAWYPGQAGATAIAEVLTGRVNPGGRLPITFPVDLHQTPRPQLPGAGL
MSMEG_5144|M.smegmatis_MC2_155 LIQAFMPGEEGGAAIAGVLSGRVQPGGKLPVQIPRTPGGQPGTYLQPPLG
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1910|M.thermoresistible__bu --------------------------------------------------
MLBr_02569|M.leprae_Br4923 --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 PWGTSTTIHYTEGADVGYRWFASTNQTPMFAFGHGLSYTSFEYRDLVVTG
Rv0186|M.tuberculosis_H37Rv PWGTSTTIHYTEGADVGYRWFASTNQTPMFAFGHGLSYTSFEYRDLVVTG
MMAR_0430|M.marinum_M PVGAPMRIHYREGADIGYRWFATTGEVPNFAFGHGLTYTSFGYRDLAVSG
MUL_1080|M.ulcerans_Agy99 PVGAPMRIHYREGADIGYRWFATTGEVPNFAFGHGLTYTSFGYRDLAVSG
MAV_4994|M.avium_104 PWGTPSTIDYVEGADVGYRWFAAKGHVPMFAFGHGLSYTRFEHRDLVVRG
MSMEG_5144|M.smegmatis_MC2_155 TLG---------------ADVSSVDPTPLYPFGYGRSYTTFEVDDLRISS
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1910|M.thermoresistible__bu --------------------------------------------------
MLBr_02569|M.leprae_Br4923 --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 GHT-----VHASFSVTNTGDRSGADVPQLYMIA---APGESRLRLLGFER
Rv0186|M.tuberculosis_H37Rv GHT-----VHASFSVTNTGDRSGADVPQLYMIA---APGESRLRLLGFER
MMAR_0430|M.marinum_M GDT-----VTVTFGVANTGDRSGADVPQVYLTA---VTGEPRLRLLGFER
MUL_1080|M.ulcerans_Agy99 GDT-----VTVTFGVANTGDRSGADVPQVYLTA---VTGEPRLRLLGFER
MAV_4994|M.avium_104 GDT-----ITAGFTVVNTGDRAGADVPQLYLTA---MPGKPCLRLLGFER
MSMEG_5144|M.smegmatis_MC2_155 ERVPTDGDWTVSVRVRNTGPRDGDEVVQLYLEDPVASVARPTRQLMGYAR
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1910|M.thermoresistible__bu --------------------------------------------------
MLBr_02569|M.leprae_Br4923 --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 VELEPGQTRRVRIEADPRLLARYDGEARSWRIEPGGYTVAVGASAVALKL
Rv0186|M.tuberculosis_H37Rv VELEPGQTRRVRIEADPRLLARYDGEARSWRIEPGGYTVAVGASAVALKL
MMAR_0430|M.marinum_M VELEPGQSCAVEIVADPRLLACYDGAAGCWRIEAGTYTVAVGASAAALSA
MUL_1080|M.ulcerans_Agy99 VELEPGQSCAVEIVADPRLLACYDGAAGCWRIEAGTYTVAVGASAAALSA
MAV_4994|M.avium_104 VELGPGESRHVTIDADPRLLAHYDGSIRSWRIAAGSHTVALGISATTLRS
MSMEG_5144|M.smegmatis_MC2_155 VTVPAASAVTVTFGLHADR-TAFTGRDLTRIVEAGQVDLLVGTSAADIAC
Mflv_0415|M.gilvum_PYR-GCK -------------------------------
Mvan_0256|M.vanbaalenii_PYR-1 -------------------------------
TH_1910|M.thermoresistible__bu -------------------------------
MLBr_02569|M.leprae_Br4923 -------------------------------
MAB_4458c|M.abscessus_ATCC_199 -------------------------------
Mb0192|M.bovis_AF2122/97 AAKVKLAGRGFGR------------------
Rv0186|M.tuberculosis_H37Rv AAKVKLAGRGFGR------------------
MMAR_0430|M.marinum_M VAEVELAGRTFGR------------------
MUL_1080|M.ulcerans_Agy99 VAEAELAGRTFGR------------------
MAV_4994|M.avium_104 TATVELTARTFGR------------------
MSMEG_5144|M.smegmatis_MC2_155 RGTVRLTGPTRTVGSDRTMLTPVTVVPEDPR