For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDPLAHAPTLTLTQQAALGSGASFWTTEAVGHIPSIVLTDGPHGVRRQSGASDHLGVGGSEPATCFPPAV ALSQTWDPDLVERVGAALGAEARALGVHVLLGPGVNIKRDPRCGRNFEYFSEDPLLSGVLGAAWVRGVQG RGVGASLKHFAVNNSEHDRMRASSDVDARTLREIYLRAFEHVVREAAPWTVMCSNNRINGVYASENTWLL TDVLRGEWGFDGVVVSDWGAVRDRVAAVAAGLDLEMPSSGGFGDAEVVAAVTAGALDARAVTTAAERVAG LAVRAYESHSAAGHFDIGELTEAHHRLACEAAAQAVVLLKNDLRDGEPILPLAPSRLAVIGPFAEEPRHQ GGGSSHVNPTRLDIPLDEIRARNGDQAATHFPGLPTDSGQDSEQVRAEAVRGAAEAEAAVLFLGLTADEE SEGYDREHIDLPQAQLALLRAVVEVQPRTVVVLSHGGVVRLDEVAKHAPAILDGALLGQAGGGAVADVLF GAVNPSGRLAETVPVRLQDCPAFLNFPSENGHTVYGERVFVGYRWYDARALEVTFPFGHGLSYTAFAFSD LEVDSDGPTVRVVVANVGPRAGREVVQVYVSKPDSQISRPPRWLAGFGSLSLESGERRTLEIPIRRTDLA FWEIRSGQWGVEPGDYTVSVGASSRDLRLHATVTVEGDVPRAVFTRESTLGEILADPAAAPLLFETLGTA AQDAPAMGTDLLRMLESVPIERMVALSGGKVTPERLDDLLAALNAVRP*
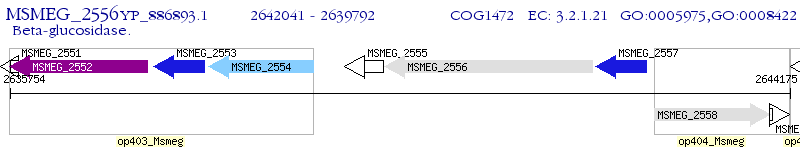
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2556 | - | - | 100% (749) | beta-glucosidase |
| M. smegmatis MC2 155 | MSMEG_5144 | - | 2e-70 | 33.23% (662) | xylosidase/arabinosidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0192 | bglS | 2e-85 | 35.31% (674) | beta-glucosidase BGLS |
| M. gilvum PYR-GCK | Mflv_0415 | - | 6e-06 | 28.03% (239) | Beta-N-acetylhexosaminidase |
| M. tuberculosis H37Rv | Rv0186 | bglS | 2e-85 | 35.31% (674) | beta-glucosidase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4458c | - | 3e-06 | 26.04% (288) | putative secreted hydrolase |
| M. marinum M | MMAR_0430 | bglS | 5e-84 | 34.41% (680) | beta-glucosidase BglS |
| M. avium 104 | MAV_4994 | - | 4e-82 | 35.48% (668) | glycosyl hydrolase family protein 3 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1080 | bglS | 1e-83 | 34.26% (680) | beta-glucosidase BglS |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0192|M.bovis_AF2122/97 --------------------MTDDERFSLLVGLTGASDLWPVRDERIPQG
Rv0186|M.tuberculosis_H37Rv --------------------MTDDERFSLLVGLTGASDLWPVRDERIPQG
MMAR_0430|M.marinum_M MSSSGHNAPADVRARQVESEMTDEERFSLLLGVMGAGDLWPLRDERIPAD
MUL_1080|M.ulcerans_Agy99 --------------------MTDEERFSLLLGVMGAGDLWPLRDERIPAD
MAV_4994|M.avium_104 ------MKDPDERARTVEAQMTDDERFSLLVAVMGAGDMWPVRDERIPAD
MSMEG_2556|M.smegmatis_MC2_155 --------------------MTDPLAHAPTLTLTQQAALGSGASFWTTEA
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 ------------------------------------MRAGTVLTGLVVAA
Mb0192|M.bovis_AF2122/97 VPMCAGYVPGIPRLGVPALLMSDAGLGVTNPGYRPGDTATALPAGLALAA
Rv0186|M.tuberculosis_H37Rv VPMCAGYVPGIPRLGVPALLMSDAGLGVTNPGYRPGDTATALPAGLALAA
MMAR_0430|M.marinum_M VPMSAGYVPGIARLGVPALLMSDAGLGVTNPGYRPGDTATALPAGLALAA
MUL_1080|M.ulcerans_Agy99 VPMSAGYVPGIARLGVPALLMTDAGLGVTNPGYRPGDTATALPAGLALAA
MAV_4994|M.avium_104 VPMSAGYVPGVPRLGVPPLLMSDAGLGVTNPGYRPGDTATALPAGLALAA
MSMEG_2556|M.smegmatis_MC2_155 VGHIPSIVLTDGPHGVRRQSGASDHLGVGGS-----EPATCFPPAVALSQ
Mflv_0415|M.gilvum_PYR-GCK -----------------------------------------------MST
MAB_4458c|M.abscessus_ATCC_199 LTACGGNASEPSKENTSSTKAQAP------------ATAAPAADLACLST
::
Mb0192|M.bovis_AF2122/97 SFNPVLARSSGKAIGREARSRGFNVQLAGAINLARDPRNGRNFEYLSEDP
Rv0186|M.tuberculosis_H37Rv SFNPVLARSSGKAIGREARSRGFNVQLAGAINLARDPRNGRNFEYLSEDP
MMAR_0430|M.marinum_M SFNPALARSAGEVIGREARSRGFNVQLAGAMNLARDPRNGRNFEYLSEDP
MUL_1080|M.ulcerans_Agy99 SFNPALARSAGEVIGREARSRGFNVQLAGAMNLARDPRNGRNFEYLSEDP
MAV_4994|M.avium_104 TFDPALARAAGELIGREARSRGFNVQLAGAMNLARDPRNGRNFEYLSEDP
MSMEG_2556|M.smegmatis_MC2_155 TWDPDLVERVGAALGAEARALGVHVLLGPGVNIKRDPRCGRNFEYFSEDP
Mflv_0415|M.gilvum_PYR-GCK RDKLAQLLMVGVTGAADARTAAAEYHVGGIMVG-----SWTDLSMLTDGS
MAB_4458c|M.abscessus_ATCC_199 RDRLAQLLMVGIKDADDARALVRDHHVGGIFVG-----SWTKKEMLTDGS
* . :**: . :. . . . :::..
Mb0192|M.bovis_AF2122/97 LLSATMAAESIIGIQQQGVIATTKHFSLNCNETNRHWLDAVIDPDAHRES
Rv0186|M.tuberculosis_H37Rv LLSATMAAESIIGIQQQGVIATTKHFSLNCNETNRHWLDAVIDPDAHRES
MMAR_0430|M.marinum_M FLTAAMVAESIIGIQQQGVISTVKHFSLNCNETNRHWLDAVIDPDAHRES
MUL_1080|M.ulcerans_Agy99 FLTAAMVAESIIGIQQQGVISTVKHFSLNCNETNRHWLDAVIDPDAHRES
MAV_4994|M.avium_104 LLTAAMVAESVAGIQQQGVISTVKHYSLNCNETNRHFLDAVIDPDAHRES
MSMEG_2556|M.smegmatis_MC2_155 LLSGVLGAAWVRGVQGRGVGASLKHFAVNNSEHDRMRASSDVDARTLREI
Mflv_0415|M.gilvum_PYR-GCK LPDIANAGLLPLAVS--------------------------VDEEGGRVS
MAB_4458c|M.abscessus_ATCC_199 LHDVITAGPIPAAVS--------------------------IDEEGGRVS
: . .:. :* *
Mb0192|M.bovis_AF2122/97 DLLAFEIVIERSQPGAVMAAYNKVNGDYAAGNDHLLNDVLKGAWGYRGWV
Rv0186|M.tuberculosis_H37Rv DLLAFEIVIERSQPGAVMAAYNKVNGDYAAGNDHLLNDVLKGAWGYRGWV
MMAR_0430|M.marinum_M DLLAFEIAIERAQPGAVMAAYNKVNGVYAAANGELINEVLKGAWGYRGWV
MUL_1080|M.ulcerans_Agy99 DLLAFEIAIERAQPGAVVAAYNKVNGVYATANGELINEVLKGAWGYRGWV
MAV_4994|M.avium_104 DLLAFELAIERAQPGAVMTAYNKVNGVYAAANSVLLNDVLKDAWAYRGWV
MSMEG_2556|M.smegmatis_MC2_155 YLRAFEHVVREAAPWTVMCSNNRINGVYASENTWLLTDVLRGEWGFDGVV
Mflv_0415|M.gilvum_PYR-GCK RLSGLIGPQPSARVLAQTKTPDEVRAIARDRGAKMRNLGITVDFAPVVDV
MAB_4458c|M.abscessus_ATCC_199 RLKDLIGPAPSARELAATQTPEQVQALSQERAQKMKDLGITIDFAPDADV
* : : : : :.:.. : : :. *
Mb0192|M.bovis_AF2122/97 MSDWGGTPSWECALAGLDQECGAQIDAVLWQSEAFTDRLRAAYADGNLPK
Rv0186|M.tuberculosis_H37Rv MSDWGGTPSWECALAGLDQECGAQIDAVLWQSEAFTDRLRAAYADGNLPK
MMAR_0430|M.marinum_M MSDWGGTPGWECALGGLDQECGAQIDAILWQSEAFGDPLRAAYHEGRLPK
MUL_1080|M.ulcerans_Agy99 MSDWGGTPGWECALGGLDQECGAQIDAILWQSEAFGDPLRAAYHEGRLPK
MAV_4994|M.avium_104 MSDWGATPGWECALGGLDQECGAQIDALLWQAESFGAPLRDAYADGRLPK
MSMEG_2556|M.smegmatis_MC2_155 VSDWGAVRDRVAAVA-------AGLDLEMPSSGGFGDAEVVAAVTAGALD
Mflv_0415|M.gilvum_PYR-GCK SD--------------------APDNTVIGDRSFGADPTTVTDYAGAYAQ
MAB_4458c|M.abscessus_ATCC_199 TD--------------------GDPDGAIGDRSFSGDPQKVAEYAEASVR
. . : : . :
Mb0192|M.bovis_AF2122/97 GRLSDMVRRILRSMFAVGIDRWKPAPAPDM----NAHNEIAAQMARQGIV
Rv0186|M.tuberculosis_H37Rv GRLSDMVRRILRSMFAVGIDRWKPAPAPDM----NAHNEIAAQMARQGIV
MMAR_0430|M.marinum_M ARLSEMVRRILRSMFAVGVDRWEPAPAPDM----DAHDEVAAEIARQGIV
MUL_1080|M.ulcerans_Agy99 ARLSEMVRRILRSMFAVGVDRWEPAPAPDM----DAHDEVAAEIARQGIV
MAV_4994|M.avium_104 DRLSDMVRRILRSIFAVGVDRSDRAPAPDL----NAHNDIALRIARQGIV
MSMEG_2556|M.smegmatis_MC2_155 ARAVTTAAERVAGLAVRAYESHSAAGHFDIGELTEAHHRLACEAAAQAVV
Mflv_0415|M.gilvum_PYR-GCK GLRDAGVLPVLKHFPGHGS---------------GSGDSHVGSVVAPSLE
MAB_4458c|M.abscessus_ATCC_199 GYQAGGVRAVVKHFPGHGH---------------ASGDSHTNGVVTPPLE
. : : . : . . . :
Mb0192|M.bovis_AF2122/97 LLQN--RGLLPLAPESAGR-IAVIGGYAHLGVPAGYGSSAVTPPGGYAGV
Rv0186|M.tuberculosis_H37Rv LLQN--RGLLPLAPESAGR-IAVIGGYAHLGVPAGYGSSAVTPPGGYAGV
MMAR_0430|M.marinum_M LLQN--RGVLPLTPAPGTR-IAVIGGYAQLGVVAGYGSSAVVPPGGYAEV
MUL_1080|M.ulcerans_Agy99 LLQN--RGVLPLRPAPGTR-IAVIGGYAQLGVLAGYGSSAVVPPGGYAEV
MAV_4994|M.avium_104 LLTN--RGLLPLRPDSPAR-IAVIGGYAQLGVPAGFGSSTVVPAGGYAAV
MSMEG_2556|M.smegmatis_MC2_155 LLKNDLRDGEPILPLAPSR-LAVIGPFAEEPRHQGGGSSHVNPTRLDIPL
Mflv_0415|M.gilvum_PYR-GCK QLQA--SDLVPYRTLAAQAPVGVMVGHVEVPGLTGSDPASLSP-------
MAB_4458c|M.abscessus_ATCC_199 QLKD--NDLVPYRSLVTSG-AAVMIGHMQVPGLTGDDPASLSG-------
* . * . .*: . . * ..: :
Mb0192|M.bovis_AF2122/97 IPIGGSGLAAGLRNLYLLPSSPLSELRKRLPNAQFEFDPGINPAEAVLAA
Rv0186|M.tuberculosis_H37Rv IPIGGSGLAAGLRNLYLLPSSPLSELRKRLPNAQFEFDPGINPAEAVLAA
MMAR_0430|M.marinum_M IPIGGAGLTAGVRNLYLLGSSPLDELRTQFGNAELEFDPGINPAEAVAVA
MUL_1080|M.ulcerans_Agy99 IPIGGAGLTPGVRNLYLLGSSPLDELRTQFGNAELEFDPGINPAEAVAVA
MAV_4994|M.avium_104 VPIGGTALEAGLRNLYLLPSSPLQELRKLLPENTIDFDPGISPAEAVAAA
MSMEG_2556|M.smegmatis_MC2_155 DEIRARNGDQAATHFPGLPTDSGQDSEQVR----------AEAVRGAAEA
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 RRADIAIVFAIRAEGEGFDSADLSLPWGQDALIAAVASANANTVVVLETG
Rv0186|M.tuberculosis_H37Rv RRADIAIVFAIRAEGEGFDSADLSLPWGQDALIAAVASANANTVVVLETG
MMAR_0430|M.marinum_M RRADVAVVFAIRAEGEGFDSADLTLPWGQDAVISAVAAANPNTVVVLETG
MUL_1080|M.ulcerans_Agy99 RRADVAVVFAIRAEGEGFDSADLTLPWGQDAVISAVAAANPNTVVVLEGG
MAV_4994|M.avium_104 QRADIAIVFAVRAEGEGFDLADLSLPSGQDELIGAVAAANPNTVVVLQTG
MSMEG_2556|M.smegmatis_MC2_155 EAAVLFLGLTADEESEGYDREHIDLPQAQLALLRAVVEVQPRTVVVLSHG
Mflv_0415|M.gilvum_PYR-GCK -QAYGLLRSG-GYGGPPFGGVVFTDDLSGMQAISDRLGVAEAVLRALQAG
MAB_4458c|M.abscessus_ATCC_199 -AAVDLLRKGTGYGAPAYDGVIFTDDLSSMAAITDRFPIEEAVLKSIKAG
* : . :. : . : .: :. *
Mb0192|M.bovis_AF2122/97 NPVTMPWRDSVN-AIMQAWYPGQAGGQAVAEIVTGQVNPSGRLPITFPVD
Rv0186|M.tuberculosis_H37Rv NPVTMPWRDSVN-AIMQAWYPGQAGGQAVAEIVTGQVNPSGRLPITFPVD
MMAR_0430|M.marinum_M NPVAMPWQDSVN-AIVQAWYPGQAGARVIAEVLAGRVNPCGRLPITFPVG
MUL_1080|M.ulcerans_Agy99 NPVAMPWQDSVN-AIVQAWYPGQAGARVIAEVLAGRVNPCGRLPITFPVG
MAV_4994|M.avium_104 NPVTMPWLGAVN-AVVQAWYPGQAGATAIAEVLTGRVNPGGRLPITFPVD
MSMEG_2556|M.smegmatis_MC2_155 GVVRLDEVAKHAPAILDGALLGQAGGGAVADVLFGAVNPSGRLAETVPVR
Mflv_0415|M.gilvum_PYR-GCK ADVALWLSTGEVPAVLDRLEKAVGAGELTMSGVDGAVTRIVAMKGPSPRC
MAB_4458c|M.abscessus_ATCC_199 VDWALWVSTEKVGSVLDRLESAYNAGELNADAINASVRRVLAYKG-VPAC
: :::: . .. . : . * *
Mb0192|M.bovis_AF2122/97 LGQTPRSQPRELGAPWGTSTTIHYTEGADVGYRWFASTNQTPMFAFGHGL
Rv0186|M.tuberculosis_H37Rv LGQTPRSQPPELGAPWGTSTTIHYTEGADVGYRWFASTNQTPMFAFGHGL
MMAR_0430|M.marinum_M LGQTPRPELPGYGAPVGAPMRIHYREGADIGYRWFATTGEVPNFAFGHGL
MUL_1080|M.ulcerans_Agy99 LGQTPRPELPGYGAPVGAPMRIHYREGADIGYRWFATTGEVPNFAFGHGL
MAV_4994|M.avium_104 LHQTPRPQLPGAGLPWGTPSTIDYVEGADVGYRWFAAKGHVPMFAFGHGL
MSMEG_2556|M.smegmatis_MC2_155 LQDCP-----AFLNFPSENGHTVYGERVFVGYRWYDARALEVTFPFGHGL
Mflv_0415|M.gilvum_PYR-GCK GG------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 G-------------------------------------------------
Mb0192|M.bovis_AF2122/97 SYTSFEYRDLVVTGGHTVHASFSVTNTGDRSGADVPQLYMIAAPG---ES
Rv0186|M.tuberculosis_H37Rv SYTSFEYRDLVVTGGHTVHASFSVTNTGDRSGADVPQLYMIAAPG---ES
MMAR_0430|M.marinum_M TYTSFGYRDLAVSGGDTVTVTFGVANTGDRSGADVPQVYLTAVTG---EP
MUL_1080|M.ulcerans_Agy99 TYTSFGYRDLAVSGGDTVTVTFGVANTGDRSGADVPQVYLTAVTG---EP
MAV_4994|M.avium_104 SYTRFEHRDLVVRGGDTITAGFTVVNTGDRAGADVPQLYLTAMPG---KP
MSMEG_2556|M.smegmatis_MC2_155 SYTAFAFSDLEVDS-DGPTVRVVVANVGPRAGREVVQVYVSKPDSQISRP
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 RLRLLGFERVELEPGQTRRVRIEADPRLLARYDGEARSWRIEPGGYTVAV
Rv0186|M.tuberculosis_H37Rv RLRLLGFERVELEPGQTRRVRIEADPRLLARYDGEARSWRIEPGGYTVAV
MMAR_0430|M.marinum_M RLRLLGFERVELEPGQSCAVEIVADPRLLACYDGAAGCWRIEAGTYTVAV
MUL_1080|M.ulcerans_Agy99 RLRLLGFERVELEPGQSCAVEIVADPRLLACYDGAAGCWRIEAGTYTVAV
MAV_4994|M.avium_104 CLRLLGFERVELGPGESRHVTIDADPRLLAHYDGSIRSWRIAAGSHTVAL
MSMEG_2556|M.smegmatis_MC2_155 PRWLAGFGSLSLESGERRTLEIPIRRTDLAFWEIRSGQWGVEPGDYTVSV
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 GASAVALKLAAKVKLAGRGFGR----------------------------
Rv0186|M.tuberculosis_H37Rv GASAVALKLAAKVKLAGRGFGR----------------------------
MMAR_0430|M.marinum_M GASAAALSAVAEVELAGRTFGR----------------------------
MUL_1080|M.ulcerans_Agy99 GASAAALSAVAEAELAGRTFGR----------------------------
MAV_4994|M.avium_104 GISATTLRSTATVELTARTFGR----------------------------
MSMEG_2556|M.smegmatis_MC2_155 GASSRDLRLHATVTVEGDVPRAVFTRESTLGEILADPAAAPLLFETLGTA
Mflv_0415|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0192|M.bovis_AF2122/97 ------------------------------------------------
Rv0186|M.tuberculosis_H37Rv ------------------------------------------------
MMAR_0430|M.marinum_M ------------------------------------------------
MUL_1080|M.ulcerans_Agy99 ------------------------------------------------
MAV_4994|M.avium_104 ------------------------------------------------
MSMEG_2556|M.smegmatis_MC2_155 AQDAPAMGTDLLRMLESVPIERMVALSGGKVTPERLDDLLAALNAVRP
Mflv_0415|M.gilvum_PYR-GCK ------------------------------------------------
MAB_4458c|M.abscessus_ATCC_199 ------------------------------------------------