For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFRSASRPLGLRAAIAALAFTALPGAIILLAPAGTAAADVCASAGRRITVGGCVNVADAVAPYVPPPAYY APLPGEAPPPPPPPPPGSNVTGCVGWNGRWVSASGCN
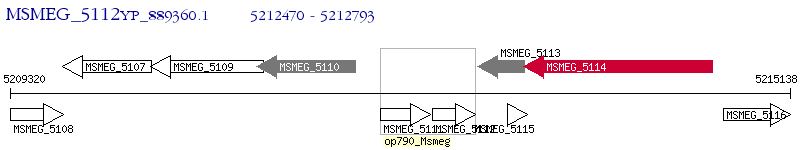
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5112 | - | - | 100% (107) | hypothetical protein MSMEG_5112 |
| M. smegmatis MC2 155 | MSMEG_5794 | - | 8e-06 | 32.97% (91) | hypothetical protein MSMEG_5794 |
| M. smegmatis MC2 155 | MSMEG_4234 | - | 2e-05 | 72.73% (22) | hypothetical protein MSMEG_4234 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0262c | PPE2 | 4e-05 | 36.11% (72) | PPE family protein |
| M. gilvum PYR-GCK | Mflv_3145 | - | 4e-21 | 50.00% (96) | hypothetical protein Mflv_3145 |
| M. tuberculosis H37Rv | Rv0256c | PPE2 | 4e-05 | 36.11% (72) | PPE family protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4000 | - | 9e-26 | 55.67% (97) | hypothetical protein MAB_4000 |
| M. marinum M | MMAR_2607 | - | 2e-31 | 64.89% (94) | hypothetical protein MMAR_2607 |
| M. avium 104 | MAV_2236 | - | 9e-18 | 50.00% (76) | hypothetical protein MAV_2236 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1502 | - | 4e-05 | 40.91% (44) | two component regulator - sensor kinase |
| M. vanbaalenii PYR-1 | Mvan_1623 | - | 9e-06 | 36.05% (86) | hypothetical protein Mvan_1623 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0262c|M.bovis_AF2122/97 MTAPIWMASPPEVHSALLSSGPGPGPLLVSAEGWHSLSIAYAETADELAA
Rv0256c|M.tuberculosis_H37Rv MTAPIWMASPPEVHSALLSSGPGPGPLLVSAEGWHSLSIAYAETADELAA
MSMEG_5112|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4000|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3145|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2607|M.marinum_M --------------------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 ----------------------------------------------MAEE
Mb0262c|M.bovis_AF2122/97 LLAAVQAGTWDGPTAAVYVAAHTPYLAWLVQASANSAAMATRQETAATAY
Rv0256c|M.tuberculosis_H37Rv LLAAVQAGTWDGPTAAVYVAAHTPYLAWLVQASANSAAMATRQETAATAY
MSMEG_5112|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4000|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3145|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2607|M.marinum_M --------------------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 LESPGDPETPTPSLQRRVILLVVALLAVLLVVLGVTIDVSLGLQARRNLH
Mb0262c|M.bovis_AF2122/97 GTALAAMPTLAELGANHALHGVLMATNFFGINTIPIALNESDYARMWIQA
Rv0256c|M.tuberculosis_H37Rv GTALAAMPTLAELGANHALHGVLMATNFFGINTIPIALNESDYARMWIQA
MSMEG_5112|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4000|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3145|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2607|M.marinum_M --------------------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 DLLLAATSRADALASVGTSPELIAAEINGGSVRALLVTADGATYGDPNIS
Mb0262c|M.bovis_AF2122/97 ATTMASYQAVSTAAVAAAPQTTPAPQIVKANAPTAASDEPNQVQEWLQWL
Rv0256c|M.tuberculosis_H37Rv ATTMASYQAVSTAAVAAAPQTTPAPQIVKANAPTAASDEPNQVQEWLQWL
MSMEG_5112|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4000|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3145|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2607|M.marinum_M --------------------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 PDTTAGPAVPALPGPPPLIPPGGHQGPHGPPPPPPPPPGGPPPGPPPDAT
Mb0262c|M.bovis_AF2122/97 QKIGYTDFYNNVIQPFINWLTNLPFLQAMFSGFDPWLPSLGNPLTFLSPA
Rv0256c|M.tuberculosis_H37Rv QKIGYTDFYNNVIQPFINWLTNLPFLQAMFSGFDPWLPSLGNPLTFLSPA
MSMEG_5112|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4000|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3145|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2607|M.marinum_M --------------------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 ATAAVHTLPDGSRLILVADTTQTTQVTRQLRQLMIGAGLVTLVVAALLLV
Mb0262c|M.bovis_AF2122/97 NIAFALGYPMDIGSYVAFLSQTFAFIGADLAAAFASGNPATIAFTLMFTT
Rv0256c|M.tuberculosis_H37Rv NIAFALGYPMDIGSYVAFLSQTFAFIGADLAAAFASGNPATIAFTLMFTT
MSMEG_5112|M.smegmatis_MC2_155 ---------------------------------MFRSASRPLGLRAAIAA
MAB_4000|M.abscessus_ATCC_1997 ----------------------------------MRRLTAALTVAVALAG
Mflv_3145|M.gilvum_PYR-GCK ---------------------------------MWT---ARVLATVG---
Mvan_1623|M.vanbaalenii_PYR-1 ---------------------------------MSQSPVERVRPTASRID
MMAR_2607|M.marinum_M ---------------------------------MGSESNEALAKEISMSP
MAV_2236|M.avium_104 ---------------------------------MNARRCRAALLVLCG--
MUL_1502|M.ulcerans_Agy99 LVSRAALAPLDRLAALAKDITTGDRGRRLRPSRGDTELGRAASAFDGMLD
Mb0262c|M.bovis_AF2122/97 VEAIG-TIITDTIALVKTLLEQTLALLPAALPLLAAPLAPLTLAPASAAG
Rv0256c|M.tuberculosis_H37Rv VEAIG-TIITDTIALVKTLLEQTLALLPAALPLLAAPLAPLTLAPASAAG
MSMEG_5112|M.smegmatis_MC2_155 -------------------------LAFTALPGAIILLAPAGTAAADVCA
MAB_4000|M.abscessus_ATCC_1997 -------------------------LPIT---------QPA--ANADVCV
Mflv_3145|M.gilvum_PYR-GCK ---------------------------LSLMTLGLSLFPQTPEASADVCG
Mvan_1623|M.vanbaalenii_PYR-1 CHNHHDLGGEGRRPIRGVALRKSWLVASAAVVLGASVPALTPAPAYAVCG
MMAR_2607|M.marinum_M WK-------------RPSATAAIAALAFTALPQVLAVMAPSGAAQADVCV
MAV_2236|M.avium_104 ---------------------------LAAVPAILVAVPGADRADATVCV
MUL_1502|M.ulcerans_Agy99 ALEDSELRAQQAATAAQRAENATRQFLVDAAHELRTPIAGMQVAAEQLAS
. .
Mb0262c|M.bovis_AF2122/97 GFAGLSG--------LAGLVGIPPSAPPVIPPVAAIAPS-----IPTPTP
Rv0256c|M.tuberculosis_H37Rv GFAGLSG--------LAGLVGIPPSAPPVIPPVAAIAPS-----IPTPTP
MSMEG_5112|M.smegmatis_MC2_155 S-AGRR-------------ITVGGCVN----VADAVAPY-----VPPP--
MAB_4000|M.abscessus_ATCC_1997 G-GGRR-------------ITVSGCAN----IADTVQRY-----APPP--
Mflv_3145|M.gilvum_PYR-GCK SIGGRH-------------VSVNACGN----VADAIAPW-----VPPP--
Mvan_1623|M.vanbaalenii_PYR-1 SISGVN-------------VDVTGCADPLYELNAAPPPP-----PPPP--
MMAR_2607|M.marinum_M N-AGRR-------------VSVSGCAN----IADAIAPY-----VPPP--
MAV_2236|M.avium_104 G-AGRR-------------VTVSGCTN----IGDNIARY-----APPP--
MUL_1502|M.ulcerans_Agy99 SAGQQQEDPAARGQYRRASLLLSDARRAGRLVADMLDLSRIDAGLPLDRH
. . : : . *
Mb0262c|M.bovis_AF2122/97 TPAPAPAPTAVTAPTPPPGPPPPPVTA---------PPPVTGAGIQSFGY
Rv0256c|M.tuberculosis_H37Rv TPAPAPAPTAVTAPTPPPGPPPPPVTA---------PPPVTGAGIQSFGY
MSMEG_5112|M.smegmatis_MC2_155 -AYYAPLPGEA--------PPPPP------------PPPPG-SNVTGCVG
MAB_4000|M.abscessus_ATCC_1997 -EDYAPLP-DD--------PPPPP------------PAP----NVHACVG
Mflv_3145|M.gilvum_PYR-GCK -AAYAPLPEDYNA-----APPPPPPPP---------PP----PNVSVCAN
Mvan_1623|M.vanbaalenii_PYR-1 -PPPPPPPPPVYV-----PPPPPPVYV---------PPVYV-PHVSVCAD
MMAR_2607|M.marinum_M -AEYAPLPEDY--------PPPPP------------PP----PNVNVCAN
MAV_2236|M.avium_104 -AVYAPLPEDDTS-----TPPPPP------------PP------------
MUL_1502|M.ulcerans_Agy99 DTDLAEIVDAESQRTTMLAPQITVTRSGLGTLTVAADPTRVAQILSNLLD
. * . .
Mb0262c|M.bovis_AF2122/97 LVGDLNSAAQARKAVGTGVRKKTPEPDSAEAPAAAAAPEEQVQPQRRRRP
Rv0256c|M.tuberculosis_H37Rv LVGDLNSAAQARKAVGTGVRKKTPEPDSAEAPAAAAAPEEQVQPQRRRRP
MSMEG_5112|M.smegmatis_MC2_155 WNGRWVSASGCN--------------------------------------
MAB_4000|M.abscessus_ATCC_1997 YNGRWVSASGCNP-------------------------------------
Mflv_3145|M.gilvum_PYR-GCK -VGRRISVSGCI--------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 -VGRRISVAGCV--------------------------------------
MMAR_2607|M.marinum_M -VGRRISVSGCV--------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 NARRYTPAGGAIAIDLRAGQGVATVTVTDTGPGIPDDERNRIFERLVRLD
Mb0262c|M.bovis_AF2122/97 KIKQLGRGYEYLDLDPETGHDPTGSPQGAGTLGFAGTTHKASPGQVAGLI
Rv0256c|M.tuberculosis_H37Rv KIKQLGRGYEYLDLDPETGHDPTGSPQGAGTLGFAGTTHKASPGQVAGLI
MSMEG_5112|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4000|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3145|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1623|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2607|M.marinum_M --------------------------------------------------
MAV_2236|M.avium_104 --------------------------------------------------
MUL_1502|M.ulcerans_Agy99 AARTRDHGGAGLGLPIARALARAHGGELVCLPHQGGARFRLTLPLAVAGG
Mb0262c|M.bovis_AF2122/97 TLPNDAFGGSPRTPMMPGTWDTDSATRVE
Rv0256c|M.tuberculosis_H37Rv TLPNDAFGGSPRTPMMPGTWDTDSATRVE
MSMEG_5112|M.smegmatis_MC2_155 -----------------------------
MAB_4000|M.abscessus_ATCC_1997 -----------------------------
Mflv_3145|M.gilvum_PYR-GCK -----------------------------
Mvan_1623|M.vanbaalenii_PYR-1 -----------------------------
MMAR_2607|M.marinum_M -----------------------------
MAV_2236|M.avium_104 -----------------------------
MUL_1502|M.ulcerans_Agy99 VE---------------------------