For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVVCSNLSFSWPDDTVLFSDLSFSLGPGRTGLVAPNGAGKSTLLKLIAGEYRPTAGSVSVDGTVGYLPQS LPFTGDLTVAAVLGIAPVLDALTALAAGDASDDVFTTIGDDWDIEERSRAQLDRLGLDTVALDRRLDTLS GGEVVSLGLAAQLLKRPEVLMLDEPTNNLDSAARQRFYDALDGYSGCLLVVSHDRALLDRMDQIAELYRG EVVFYGGNFTAYRETVEANQRAAESSIRNAEQQLKREKRQMQEARERAARRSSAAARNLKDAGLPKIVAG KLKRDAQESAAKSDDVHAKRVDAARARLEDAERALRDDKVIVLDLPDTEVPAGRTLFTGTGLQISRGGRN LFADNGIDVSIRGPERIALTGSNGAGKSTLLRIIDGQLKPDGGTCQQADGRVAYLSQRLDLLDLDRSVAE SLAASAPSLTHTRRMHLLAQFLFRGDRIHLPVGALSGGERLRATLACVLFAEPAPQLLLLDEPTNNLDLV SVGQLESALNAYRGAFVVVSHDERFLDTIGIERRWHLEDGRLRVTAAAG*
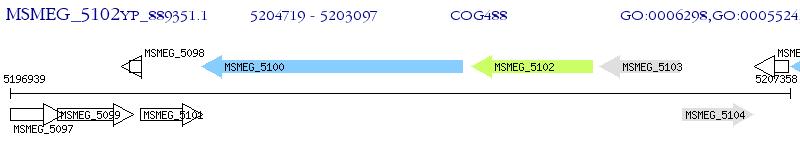
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5102 | - | - | 100% (540) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_6748 | - | 7e-61 | 33.33% (564) | ABC transporter ATPase |
| M. smegmatis MC2 155 | MSMEG_3140 | - | 5e-57 | 32.91% (550) | ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1508 | - | 3e-56 | 34.69% (539) | macrolide ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_2181 | - | 0.0 | 67.85% (535) | ABC transporter related |
| M. tuberculosis H37Rv | Rv1473 | - | 3e-56 | 34.69% (539) | macrolide ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_01248 | - | 9e-51 | 31.21% (535) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1846 | - | 1e-126 | 48.55% (519) | putative ABC transporter ATP-binding protein |
| M. marinum M | MMAR_2279 | - | 2e-55 | 33.65% (532) | ATP-binding protein ABC transporter |
| M. avium 104 | MAV_3306 | - | 7e-58 | 34.50% (542) | ABC transporter ATP-binding protein |
| M. thermoresistible (build 8) | TH_3711 | - | 0.0 | 68.67% (533) | PUTATIVE ABC transporter ATP-binding protein |
| M. ulcerans Agy99 | MUL_1481 | - | 3e-56 | 33.83% (532) | ATP-binding protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_4514 | - | 0.0 | 70.49% (532) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5102|M.smegmatis_MC2_155 ----------------------------MSVVCSNLSFSW---PDDTVLF
TH_3711|M.thermoresistible__bu ----------------------------MSIVFSDVSFAW---PDGTALF
Mflv_2181|M.gilvum_PYR-GCK MTPALPWNTCSRTEEPAPCPHLAESFRMPSITCTHLSYSFDAGPATAPLF
Mvan_4514|M.vanbaalenii_PYR-1 ---------------------------MSSVVCSHLSFSW---PDDTPLF
MAB_1846|M.abscessus_ATCC_1997 --------------------------MSAVISCGALTYTH---PDGSVAL
MMAR_2279|M.marinum_M -----------------------------MITATDLEVRAG--ARILLSP
MUL_1481|M.ulcerans_Agy99 -----------------------------MITATDVEVRAG--ARILLSP
MAV_3306|M.avium_104 -----------------------------MITATDLEVRAG--ARILLSP
Mb1508|M.bovis_AF2122/97 -----------------------------MITATDLEVRAG--ARILLAP
Rv1473|M.tuberculosis_H37Rv -----------------------------MITATDLEVRAG--ARILLAP
MLBr_01248|M.leprae_Br4923 -------------------------MAEFIYTMKKVRKAHG----DKVIL
:
MSMEG_5102|M.smegmatis_MC2_155 SDLSFSLGPG-RTGLVAPNGAGKSTLLKLIAGEYRPTAG--SVSVDGTVG
TH_3711|M.thermoresistible__bu TDLSFTIGPG-RTGLVAPNGAGKSTLLDLIAGARRPTSG--SVTVDGTVG
Mflv_2181|M.gilvum_PYR-GCK ADLTFSAGPG-RTGLVGSNGSGKSTLLRLIAGELSPTAG--AVTVDGVIG
Mvan_4514|M.vanbaalenii_PYR-1 TDLSFTVGAG-RTGLVAANGAGKSTLLRLIAGELTATTG--AVTADGVVG
MAB_1846|M.abscessus_ATCC_1997 DGLDMLVPAG-RSGLVGVNGSGKSTLLKLIAGELRPTSG--MVHASGEIG
MMAR_2279|M.marinum_M DGPDLRVQPGDRIGLVGRNGAGKTTTLRILAGESEPYAG--SVTRAGEIG
MUL_1481|M.ulcerans_Agy99 DGPDLRVQPGDRIGLVGRNGAGKTTTLRILAGESEPYAG--SVTRAGEIG
MAV_3306|M.avium_104 DGPDLRVQPGDRIGLVGRNGAGKTTTLRILAGETEPYAG--TIARSGEIG
Mb1508|M.bovis_AF2122/97 DGPDLRVQPGDRIGLVGRNGAGKTTTLRILAGEVEPYAG--SVTRAGEIG
Rv1473|M.tuberculosis_H37Rv DGPDLRVQPGDRIGLVGRNGAGKTTTLRILAGEVEPYAG--SVTRAGEIG
MLBr_01248|M.leprae_Br4923 DDVTLNFFPGAKIGVVGPNGAGKSSVLRIMAGMDKPNNGDAFLATGASVG
. : .* : *:*. **:**:: * ::** . * : . :*
MSMEG_5102|M.smegmatis_MC2_155 YLPQS-----LPFTGDLTVAAVLGIAPVLDALT-------ALAAGDASDD
TH_3711|M.thermoresistible__bu HLPQH-----LPFMPEATVGDVLGITPVLTALD-------ALAAGDASEA
Mflv_2181|M.gilvum_PYR-GCK HLPQT-----LPMRDDRRVAEVLGVAEAIDALN-------ALSDGDSRDT
Mvan_4514|M.vanbaalenii_PYR-1 YLPQT-----LPLLDDRSVADVLGVAAVIDALN-------ALAAGDPSAH
MAB_1846|M.abscessus_ATCC_1997 YLPQD-----LTIRADQSVPDLLGLTPVLAAIE-------AIENGSTDQT
MMAR_2279|M.marinum_M YLPQDPKEGDLEVLARDRVLSARGLDVLLTDLEKQQALMAEVADDAARDR
MUL_1481|M.ulcerans_Agy99 YLPQDPKEGDLEVLARDRVLSARGLDVLLTDLEKQQALMAEVADDAARDR
MAV_3306|M.avium_104 YLPQDPKEGDLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADDDARDR
Mb1508|M.bovis_AF2122/97 YLPQDPKVGDLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADEDERDR
Rv1473|M.tuberculosis_H37Rv YLPQDPKVGDLDVLARDRVLSARGLDVLLTDLEKQQALMAEVADEDERDR
MLBr_01248|M.leprae_Br4923 ILQQEPPLNEEKTVRGNVEEGLGDIKIKLDRFS----EVAELMATDSSSE
* * .: : : :
MSMEG_5102|M.smegmatis_MC2_155 VFTTIG---------DDWDIEERSRAQLDRLGLDTVALDRRLDTLSGGEV
TH_3711|M.thermoresistible__bu VFAAIG---------DDWDVEERCRGMLDRLGLGEVAVDRRLGSLSGGEV
Mflv_2181|M.gilvum_PYR-GCK VFAAIG---------DDWDVEERTRAQLDRLGLGHVALDRSLRSLSGGEV
Mvan_4514|M.vanbaalenii_PYR-1 HFAMIG---------DDWDVEERTRAQLDRLGLGHIGLDRSLRSLSGGEV
MAB_1846|M.abscessus_ATCC_1997 DFDTVG---------DDWDLAERVRAELDRLGLPAAVLDRSLGELSGGEV
MMAR_2279|M.marinum_M AIRRYGQLEERFVALGGYGAESEASRICASLGLPERVLTQQLRTLSGGQR
MUL_1481|M.ulcerans_Agy99 AIRRYGQLEERFVTLGGYGAESEASRICASLGLPERVLTQQLRTLSGGQR
MAV_3306|M.avium_104 AIRRYGQLEERFVALGGYGAESEASRICASLGLPERVLTQQLRTLSGGQR
Mb1508|M.bovis_AF2122/97 AIRRYGQLEERFVALGGYGAESEAGRICASLGLPERVLTQRLRTLSGGQR
Rv1473|M.tuberculosis_H37Rv AIRRYGQLEERFVALGGYGAESEAGRICASLGLPERVLTQRLRTLSGGQR
MLBr_01248|M.leprae_Br4923 LMEEMGRLQEELDHADAWDLDSQLEQAMDVLRCPP--PDEPVTNLSGGER
: * . :. .. * . : ****:
MSMEG_5102|M.smegmatis_MC2_155 VSLGLAAQLLKRPE----------VLMLDEPTNNLDSAARQRFYDALDGY
TH_3711|M.thermoresistible__bu VAVGLAAQLLRRPE----------VLLLDEPTNNLDADARRRLHAALDDY
Mflv_2181|M.gilvum_PYR-GCK VTLGLARELLRRPD----------VLLLDEPTNNLDADARGKLCDALDDF
Mvan_4514|M.vanbaalenii_PYR-1 VSLGLARELLRRPD----------VLLLDEPTNNLDADARHRLYDALDDF
MAB_1846|M.abscessus_ATCC_1997 IRLGLARLLLRRPD----------VLLLDEPTNNLDGESRRHLYDVVSAW
MMAR_2279|M.marinum_M RRVELARILFAASD---TGAGSSTVLLLDEPTNHLDADSVGWLRDFLRAH
MUL_1481|M.ulcerans_Agy99 RRVELARILFAASD---TGAGSSTVLLLDEPTNHLDADSVGWLRDFLRAH
MAV_3306|M.avium_104 RRVELARILFAAAEGGATGSGSATTLLLDEPTNHLDADSIGWLRDFLRAH
Mb1508|M.bovis_AF2122/97 RRVELARILFAASE---SGAGNSTTLLLDEPTNHLDADSLGWLRDFLRLH
Rv1473|M.tuberculosis_H37Rv RRVELARILFAASE---SGAGNSTTLLLDEPTNHLDADSLGWLRDFLRLH
MLBr_01248|M.leprae_Br4923 RRVALCKLLLSKPD----------LLLLDEPTNHLDAESVQWLEQHLAGY
: *. *: .: *:******:**. : : :
MSMEG_5102|M.smegmatis_MC2_155 SGCLLVVSHDRALLDRMDQ---IAELYRGEVVFYGGNFTAYRETVEANQR
TH_3711|M.thermoresistible__bu PGTLLVVSHDRVLLDRMER---IAELYRGEMLFYGGNFTTYQRQLAADQQ
Mflv_2181|M.gilvum_PYR-GCK TGCLLLVSHDRMLLDRMDR---IAELDRGEVVLYGGNFTAYQGAVADAQR
Mvan_4514|M.vanbaalenii_PYR-1 GGCLLVVSHDRVLLDRMDR---IAELYRGQVRFYGGNFTAYQDAVTDAQR
MAB_1846|M.abscessus_ATCC_1997 QRTLLVVSHDRELLEHVER---IGDLRDGGVTWYGGGYSDYAAAVAAEQE
MMAR_2279|M.marinum_M TGGLIIISHNVELLADVVNRVWFLDAVRGEVDVYNMGWHKYLDARATDEQ
MUL_1481|M.ulcerans_Agy99 TGGLIIISHNVELLADVVNRVWFLDAVRGEVDVYNMGWHKYLDARATDEQ
MAV_3306|M.avium_104 TGGLVIISHNVELLADVVNRVWFLDAVRGEVDVYNMSWQKYLDARATDEQ
Mb1508|M.bovis_AF2122/97 TGGLVVISHNVDLVADVVNKVWFLDAVRGQVDVYNMGWQRYVDARATDEQ
Rv1473|M.tuberculosis_H37Rv TGGLVVISHNVDLVADVVNKVWFLDAVRGQVDVYNMGWQRYVDARATDEQ
MLBr_01248|M.leprae_Br4923 AGAVLAVTHDRYFLDNVAE--WILELDRGRAYPYEGNYSTYLEKKAERLT
:: ::*: :: : . : : * * .: *
MSMEG_5102|M.smegmatis_MC2_155 AAESSIRNAEQQLKREKRQMQEARERAARRSSAAARNLKDAGLPKIVAGK
TH_3711|M.thermoresistible__bu SARNDVRHAEQSLRREKRERQQARERAARRSGTAARTVKDAGLPRIVAGA
Mflv_2181|M.gilvum_PYR-GCK AARDTVRNAEQTIKREQRQRQQARERADRRSGNAARTLKDAGLPKIVAGA
Mvan_4514|M.vanbaalenii_PYR-1 AAQDAVRSAEQLLRREKHQRQQARERADRRAGTAARTLGDAGLPKIVAGA
MAB_1846|M.abscessus_ATCC_1997 AAAQAISTAKAEVRRERRDLAEAERVIAQRRRYGAKMQANKREPKIVMGL
MMAR_2279|M.marinum_M --------------RRRRERANAERKATALRTQAAKMGAKATKAVAAQNM
MUL_1481|M.ulcerans_Agy99 --------------RRRRERANAERKATALRTQAAKMGAKATKAVAAQNM
MAV_3306|M.avium_104 --------------RRRRERANAERKAAALRTQAAKLGAKATKAVAAQNM
Mb1508|M.bovis_AF2122/97 --------------RRIRERANAERKAAALRAQAAKLGAKATKAVAAQNM
Rv1473|M.tuberculosis_H37Rv --------------RRIRERANAERKAAALRAQAAKLGAKATKAVAAQNM
MLBr_01248|M.leprae_Br4923 -----------------TQGRKDAKLQKRLTDELAWVRSGAKARQAKSKA
: : . *
MSMEG_5102|M.smegmatis_MC2_155 LKRDAQESAAKSDDVHAKRVDAARARLEDAERALRDDKVIVLDLPDTEVP
TH_3711|M.thermoresistible__bu MKRRAQESAGRTDEVRAARIAAGRARLEEAEKRLRDDDAMVLDLPDTTVP
Mflv_2181|M.gilvum_PYR-GCK RKRRAQESAARADDVHARRIDEAKLRLVSAERAVRDDDELALQLPATTVA
Mvan_4514|M.vanbaalenii_PYR-1 RKRRAQETAGRSDDVHASRVDDARSRLEDAERAVRDDDALVLELPGTLVP
MAB_1846|M.abscessus_ATCC_1997 RKRAAQESAAALKQTQTDRLDKARENLGEAQTRLRADRSIRIDLPGTEVP
MMAR_2279|M.marinum_M LRRADRMMAALDEERVADKVARIKF--------------------PTPAA
MUL_1481|M.ulcerans_Agy99 LRRADRMMAALDEERVADKVARIKF--------------------PTPAA
MAV_3306|M.avium_104 LRRADRMMAALDEERVADKVARIKF--------------------PTPAA
Mb1508|M.bovis_AF2122/97 LRRADRMMAALDEERVADKVARIKF--------------------PTPAA
Rv1473|M.tuberculosis_H37Rv LRRADRMMAALDEERVADKVARIKF--------------------PTPAA
MLBr_01248|M.leprae_Br4923 RLQRYEEMAAEAEKNRKFDFEEIQIP--------------------VGPR
: . *. .. . : .
MSMEG_5102|M.smegmatis_MC2_155 AGRTLFTGTGLQISRGGRNLFADNGIDVSIRGPERIALTGSNGAGKSTLL
TH_3711|M.thermoresistible__bu AGRTVFAGSGLTVGNVLR------GIDLTVRGPERIALTGANGVGKTTLL
Mflv_2181|M.gilvum_PYR-GCK AGRTVLQASALQVDRGGRTLLA--GVDLTIRGPERIALTGRNGAGKTTLL
Mvan_4514|M.vanbaalenii_PYR-1 SARMVLRLNGIRVRRNGCDVLD--GIDLTVRGPERIALTGPNGAGKTTLL
MAB_1846|M.abscessus_ATCC_1997 AGRVVLTTEQLVLRHGVP-------AHLDLRGPQRVGLVGPNGSGKTTLL
MMAR_2279|M.marinum_M CGRVPLVVKGLSKTYGSLEVFT--GVDLAIDRGSRVVVLGLNGAGKTTLL
MUL_1481|M.ulcerans_Agy99 CGRVPLVVKGLSKTYGSLEVFT--GVDLAIDRGSRVVVLGLNGAGKTTLL
MAV_3306|M.avium_104 CGRTPLTATGLSKSYGSLEVFS--GVDLAIDRGSRVVVLGLNGAGKTTLL
Mb1508|M.bovis_AF2122/97 CGRTPLVANGLGKTYGSLEVFT--GVDLAIDRGSRVVILGLNGAGKTTLL
Rv1473|M.tuberculosis_H37Rv CGRTPLVANGLGKTYGSLEVFT--GVDLAIDRGSRVVILGLNGAGKTTLL
MLBr_01248|M.leprae_Br4923 LGNMVVEVEHLDKGYGGRTLIK--DLSFTLPRNGIVGIIGPNGVGKTTLF
.. . : . : : : * ** **:**:
MSMEG_5102|M.smegmatis_MC2_155 RIIDGQLKPDGGTCQQADGR-VAYLSQRLDLLDLDRSVAESLAASAPSLT
TH_3711|M.thermoresistible__bu RVIGGRLAPDAGTVTRADGR-IAYLSQRLDLLDPGQTVLDNLAEFAPTLS
Mflv_2181|M.gilvum_PYR-GCK RTLRGDVVPDAGTVQVADGR-VAYLSQRLDLLDDDRTLSDNLAMSAPTLS
Mvan_4514|M.vanbaalenii_PYR-1 RTLLGEIEPDAGAVEVADGR-VAYLSQRLDLLDEDRSVAENLAAFAPSLT
MAB_1846|M.abscessus_ATCC_1997 HTIAGRIPAAEGSATVHVP--LGLLPQRLDLLDDSLSVADNVARRAPHAD
MMAR_2279|M.marinum_M RLLAGVEHPDTGGLEPGHGLRIGYFAQEHDTLDNDATVWENIRHAAPESG
MUL_1481|M.ulcerans_Agy99 RLLAGVEHPDTGGLEPGHGLRIGYFAQEHDTLDNDATVWENIRHAAPESG
MAV_3306|M.avium_104 RLLAGAETPDTGRIEPGHGLRIGYFAQEHDTLDNDATVWQNIRHAAPDSG
Mb1508|M.bovis_AF2122/97 RLLAGVEQPDTGVLEPGYGLRIGYFAQEHDTLDNDATVWENVRHAAPDAG
Rv1473|M.tuberculosis_H37Rv RLLAGVEQPDTGVLEPGYGLRIGYFAQEHDTLDNDATVWENVRHAAPDAG
MLBr_01248|M.leprae_Br4923 KTIVGLEQPDGGAVKIGETVKLSYVDQTRAGIDPKKIVWEVVSDGLDHIQ
: : * . * :. . * :* : : :
MSMEG_5102|M.smegmatis_MC2_155 H----TRRMHLLAQFLFRGDRIHLPVGALSGGERLRATLACVLFAEPAPQ
TH_3711|M.thermoresistible__bu V----TGRRHLLARFLFDDKRIALPIRALSGGERLRATLACVLHAEPAPQ
Mflv_2181|M.gilvum_PYR-GCK V----TRRRHLLAQFLFRGDAVDLPVRALSGGERLRATLACVLFSEPAPH
Mvan_4514|M.vanbaalenii_PYR-1 V----TRRRHLLAQFLFRGDAVERPVRVLSGGERLRATLACVLYAEPAPQ
MAB_1846|M.abscessus_ATCC_1997 V----NAVRARLARFLFRGNAADKLVGALSGGERFRATLAAVLLADPAPQ
MMAR_2279|M.marinum_M E----QELRGLLGAFMFTGPQLDQPAGTLSGGEKTRLALAGLVASTAN--
MUL_1481|M.ulcerans_Agy99 E----QELRGLLGAFMFTGPQLDQPAGTLSGGEKTRLALAGLVASTAN--
MAV_3306|M.avium_104 E----QDLRGLLGAFMFSGPQLDQPAGTLSGGEKTRLALAGLVASTAN--
Mb1508|M.bovis_AF2122/97 E----QDLRGLLGAFMFTGPQLEQPAGTLSGGEKTRLALAGLVASTAN--
Rv1473|M.tuberculosis_H37Rv E----QDLRGLLGAFMFTGPQLEQPAGTLSGGEKTRLALAGLVASTAN--
MLBr_01248|M.leprae_Br4923 VGQTEVSSRAYVSAFGFKGPDQQKLAGVLSGGERNRLNLALTLKQGGN--
:. * * . .*****: * ** :
MSMEG_5102|M.smegmatis_MC2_155 LLLLDEPTNNLDLVSVGQLESALNAYRGAFVVVSHDERFLDTIGIERRWH
TH_3711|M.thermoresistible__bu LLLLDEPTNNLDLTSIAQLESALIAYRGAFIVVSHDDRFVAAIGVNRRLR
Mflv_2181|M.gilvum_PYR-GCK LLLLDEPTNNLDLAGIAQLESALVAYRGAFIVVSHDAPFLDAIAVDRVVH
Mvan_4514|M.vanbaalenii_PYR-1 LLLLDEPTNNLDLVSVGQLESALAAYRGAFIVVSHDASFLDAIGVDRVLR
MAB_1846|M.abscessus_ATCC_1997 LLLLDEPTNNLDFASYDALVSALAEYRGAVLVASHDLGFLEDIGAAQVDW
MMAR_2279|M.marinum_M VLLLDEPTNNLDPASREQVLDALRSYQGAVVLVTHDPGAAEALDPQRVVL
MUL_1481|M.ulcerans_Agy99 VLLLDEPTNNLDPASREQVLDALRSYQGAVVLVTHDPGAAEALDPQRVVL
MAV_3306|M.avium_104 VLLLDEPTNNLDPASREQVLDALRSYTGAVVLVTHDPGAAEALDPQRVVL
Mb1508|M.bovis_AF2122/97 VLLLDEPTNNLDPASREQVLDALRSYRGAVVLVTHDPGAAAALGPQRVVL
Rv1473|M.tuberculosis_H37Rv VLLLDEPTNNLDPASREQVLDALRSYRGAVVLVTHDPGAAAALGPQRVVL
MLBr_01248|M.leprae_Br4923 LILLDEPTNDLDVETLGSLENALVNFPGCAVVISHDRWFLDRTCTHILAW
::*******:** : .** : *. :: :**
MSMEG_5102|M.smegmatis_MC2_155 LEDGRLRVTAAAG-------------------------------
TH_3711|M.thermoresistible__bu LSDGKLS-------------------------------------
Mflv_2181|M.gilvum_PYR-GCK LAGGRLAEV-----------------------------------
Mvan_4514|M.vanbaalenii_PYR-1 LADSTLAPG-----------------------------------
MAB_1846|M.abscessus_ATCC_1997 RPDPVIG-------------------------------------
MMAR_2279|M.marinum_M LPDATEDYWSDEYRDLIELA------------------------
MUL_1481|M.ulcerans_Agy99 LPDATEDYWSDEYRDLIELA------------------------
MAV_3306|M.avium_104 LPDGTEDYWSDEYRDLIELA------------------------
Mb1508|M.bovis_AF2122/97 LPDGTEDYWSDEYRDLIELA------------------------
Rv1473|M.tuberculosis_H37Rv LPDGTEDYWSDEYRDLIELA------------------------
MLBr_01248|M.leprae_Br4923 EGDDEGKWFWFEGNFGAYEENKVERLGAEAARPHRVTHRKLTRD
.