For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KISLFYEFPLPRPWSEDDEHQLFQHGLDEVELADKAGFSTVWLTEHHFLEEYCHSTAPEMFLAAASQRTE NIRLGFGVMHLPPPINHPARIAERVATLDHLSNGRVEFGTGEGSSVAELGGFNIDPADKRTQWEESLEVA IRCMVEEPFSGFKGEHVEMPARNVIPKPLQKPHPPVWVACTRPSSVQMAAQKAIGALSFAYTGPEALKER VAGYYKEFEEQGAPVTPAINPNILAIGGDLSMMVARTDEEALKRLGIGGGFFSFGIMHYYLTGMHTPGRT KVWERYEQAVADDPTLAYGPGRGAIGSPETVREFLRGYEASGVDEIILLLNPRSHEGTMESIEIMGKEIL PEFIERDEKAVKDKAERLEPVVEKVESRRQGSDAPLFDETYAFGGLPTGRGGKFTAGEIPEAMAEINEGR VKAAQAEKAAREAAG*
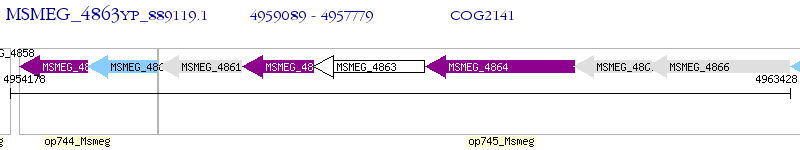
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4863 | - | - | 100% (436) | hypothetical protein MSMEG_4863 |
| M. smegmatis MC2 155 | MSMEG_6107 | - | 4e-25 | 28.74% (341) | limonene 1,2-monooxygenase |
| M. smegmatis MC2 155 | MSMEG_1475 | - | 6e-17 | 25.45% (224) | hypothetical protein MSMEG_1475 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1971 | - | 2e-20 | 27.27% (352) | monooxygenase |
| M. gilvum PYR-GCK | Mflv_2404 | - | 0.0 | 91.34% (439) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3618 | - | 6e-21 | 26.69% (341) | monooxygenase |
| M. leprae Br4923 | MLBr_00348 | - | 5e-06 | 27.49% (211) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0527c | - | 1e-26 | 28.57% (343) | monooxygenase |
| M. marinum M | MMAR_4774 | - | 0.0 | 81.99% (433) | monoxygenase |
| M. avium 104 | MAV_0881 | - | 0.0 | 85.22% (433) | hypothetical protein MAV_0881 |
| M. thermoresistible (build 8) | TH_3770 | - | 0.0 | 90.91% (429) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_0345 | - | 0.0 | 81.52% (433) | monoxygenase |
| M. vanbaalenii PYR-1 | Mvan_4242 | - | 0.0 | 92.47% (438) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3618|M.tuberculosis_H37Rv MKAP-LRFGVFITPFHPTGQSPTVALQYDMERVVALDRLGYDEAWFGEHH
MAB_0527c|M.abscessus_ATCC_199 MSAPPLRFGAFITPFHPVGQNPTTALEYDLDRVVALDRLGYHEAWFGEHH
Mflv_2404|M.gilvum_PYR-GCK MKIS--LFYEFPLPRPWSDDDEHQLFQHGLDEVELADKAGFSTVWLTEHH
Mvan_4242|M.vanbaalenii_PYR-1 MKIS--LFYEFPLPRPWSEDDEHQLFQHGLDEVELADKAGFSTVWLTEHH
MSMEG_4863|M.smegmatis_MC2_155 MKIS--LFYEFPLPRPWSEDDEHQLFQHGLDEVELADKAGFSTVWLTEHH
TH_3770|M.thermoresistible__bu MKIS--LFYEFPLPRPWSEDDEHKLFQDGLDEVEAADKAGFSSVWLTEHH
MMAR_4774|M.marinum_M MKIS--LFYEFALPRPWAPDDERIMLQDCLDEVEAADKAGFSTVWLTEHH
MUL_0345|M.ulcerans_Agy99 MKIS--LFYEFALPRPWAPNDERIMLQDCLDEVEAADKAGFSTVWLTEHH
MAV_0881|M.avium_104 MKIS--LFYEFALPRPWAPDDEHVLLQECLDEVEAADKAGFSTVWLTEHH
Mb1971|M.bovis_AF2122/97 MEIG-----IFLMPAHPPERTLYDATRWDLDVIELADQLGYVEAWVGEHF
MLBr_00348|M.leprae_Br4923 MVER----ANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAW
* : : : *: .: *
Rv3618|M.tuberculosis_H37Rv S-GGYELIACPEVFIAAAAERTTHIRLGTGVVSLP--YHHPLMVADRWVL
MAB_0527c|M.abscessus_ATCC_199 S-GGYELISCPEVFIATAAERTKHIKLGTGVVSLP--YHHPLMVVDRWIL
Mflv_2404|M.gilvum_PYR-GCK FLEEYCHSTAPELFLAAASQRTKNIRLGFGVMHLPPPINHPARIAERVAT
Mvan_4242|M.vanbaalenii_PYR-1 FLEEYCHSTAPELFLAAASQRTKNIRLGFGVMHLPPPINHPARIAERVAT
MSMEG_4863|M.smegmatis_MC2_155 FLEEYCHSTAPEMFLAAASQRTENIRLGFGVMHLPPPINHPARIAERVAT
TH_3770|M.thermoresistible__bu FLEEYCHSTAPEMFLAAASQRTENIRLGFGVMHLPPPINHPARIAERVAT
MMAR_4774|M.marinum_M FLEEYCHSTAPEIFLAAASQRTKNIRLGFGIMHLPPAVNHPARVAERIAT
MUL_0345|M.ulcerans_Agy99 FLEEYCHSTAPEIFLAAASQRTKNIRLGFGIMHLPPAVNHPARVAERIAT
MAV_0881|M.avium_104 FLEEYCHSTAPEIFLAAASQRTENIRLGFGIMHLPPAVNHPARVAERIAT
Mb1971|M.bovis_AF2122/97 T-VPWEPICAPDLLLAQALLRTQQIKLAPGAHLLP--YHHPVELAHRVAY
MLBr_00348|M.leprae_Br4923 GSDAYTP-------LAWWGSSTQRVRLGTSVVQLS--ARTPTACAMAALT
: :* * .::*. . *. . * .
Rv3618|M.tuberculosis_H37Rv LDHLTRGRVMFGTGPGALPSDAYMMGIDP--VEQRRMMQESLEAILALFR
MAB_0527c|M.abscessus_ATCC_199 LDHITRGRVIFGTGPGALPTDAYMMGIDP--VDQRRMQEESLEAILALLR
Mflv_2404|M.gilvum_PYR-GCK LDHLSNGRVEFGTGEGSSVAELGGFNIDP--ADKRAQWEEALEVSIRCMT
Mvan_4242|M.vanbaalenii_PYR-1 LDHLSNGRVEYGTGEGSSVAELGGFNIDP--ADKRTMWEEALEVSIRCMT
MSMEG_4863|M.smegmatis_MC2_155 LDHLSNGRVEFGTGEGSSVAELGGFNIDP--ADKRTQWEESLEVAIRCMV
TH_3770|M.thermoresistible__bu LDHLSNGRVEFGTGEGSSVAELGGFNIDP--ADKRTMWEEALEVSIRCMI
MMAR_4774|M.marinum_M LDLLSNGRVEFGTGESSSIGELGGFNIDP--ADKRGQWEEALEVAIRCMI
MUL_0345|M.ulcerans_Agy99 LDLLSNGRVEFGTGESSSIGELGGFNIDP--TDKRGQWEEALEVAIRCMI
MAV_0881|M.avium_104 LDLLSNGRVEFGTGESSSVGELGGFNIDP--ADKRAQWEEALEVAIRCMV
Mb1971|M.bovis_AF2122/97 FDHLAQGRFMLGVGASGIPGDWALYDVDGKNGEHREMTREALEIMLRIWT
MLBr_00348|M.leprae_Br4923 LDHLSGGRHILGLGVSGPQVVEGWYGQPFP--KPLSRTREYIDIVRQVWA
:* :: ** * * .. . . .* ::
Rv3618|M.tuberculosis_H37Rv AAPDERIDRHSDWFTLR--------EAQLHIRPYTWPY-PEIATAAMIS-
MAB_0527c|M.abscessus_ATCC_199 ATPQERINRETSWFTLR--------DAQLQLRPYTHPY-PEILTAAMFS-
Mflv_2404|M.gilvum_PYR-GCK EAPFTGFK--GEHVEMP--------ARNVIPKPLQQPH-PPVWVACTR--
Mvan_4242|M.vanbaalenii_PYR-1 EAPFTGFK--GEHVEMA--------ARNVIPKPLQQPH-PPVWVACTR--
MSMEG_4863|M.smegmatis_MC2_155 EEPFSGFK--GEHVEMP--------ARNVIPKPLQKPH-PPVWVACTR--
TH_3770|M.thermoresistible__bu EEPFSGFK--GEHVEMP--------ARNVIPKPLQKPH-PPVWVACTR--
MMAR_4774|M.marinum_M EEPFAGFT--GDHIQMP--------ARNVIPKPMQKPH-PPVWVACTR--
MUL_0345|M.ulcerans_Agy99 EEPFAGFT--GDHIQMP--------ARNVIPKPMQKPH-PPVWVACTR--
MAV_0881|M.avium_104 EEPFSGFH--GEHVQMP--------ARNVIPKPLQKPH-PPVWVACTR--
Mb1971|M.bovis_AF2122/97 EDEPWEHR--GKYWNANGIAPMFEGLMRRHIKPYQKPH-PPIGVTGFSAG
MLBr_00348|M.leprae_Br4923 REAPVVSD--GQHYPLPLTGAG-ATGLGKALKPITHPLRADIPIMLGAEG
. :* * . :
Rv3618|M.tuberculosis_H37Rv PSGPRLAGALGTSLLSLSMSVPGGYAALETAWGVVREQAAKAGRGEPDRA
MAB_0527c|M.abscessus_ATCC_199 PSGPRLAGKLGAGLLSLSASIPGGFAALENAWEVVREQAEIHGHPEPDRS
Mflv_2404|M.gilvum_PYR-GCK PSSVGMAAEKAIGALSFAYTGPEALKDRVDGYYKDFEEKGTPITPAINPN
Mvan_4242|M.vanbaalenii_PYR-1 PSSVQMAAQKAIGALSFAYTGPEALKERVDGYYKEFEEQGAPITPAINPN
MSMEG_4863|M.smegmatis_MC2_155 PSSVQMAAQKAIGALSFAYTGPEALKERVAGYYKEFEEQGAPVTPAINPN
TH_3770|M.thermoresistible__bu PSSVQMAAQKAIGALSFAYTGPGPLKERVDGYYKEFEEKGSPVTPAVNPN
MMAR_4774|M.marinum_M PATVQMAAQKCIGALSFAYTGPGPLTERVNGYYKEFEENGTPVTPAINPN
MUL_0345|M.ulcerans_Agy99 PATVQMAAQKCIGALSFAYTGPGPLTERVNGYYKEFEENGTPVTPAINPN
MAV_0881|M.avium_104 PASVQMAAQKAIGALSFAYTGPGPLTERVNGYYKEFEENGVPVTPAINPN
Mb1971|M.bovis_AF2122/97 SETLKLAGERGYIPMSLDLN-----TEYVATHWDAVEEGALRSGRTPDRR
MLBr_00348|M.leprae_Br4923 PKNVALAAEICDGWLPIFFS-----PRMADMYNEWLDEGFARTGARRSRE
. :*. :.: . :: .
Rv3618|M.tuberculosis_H37Rv DWRVLSIMHLSDSRDQAIDDCTYG-LPDFSRYFGAAGFVPLANTVEGTQ-
MAB_0527c|M.abscessus_ATCC_199 SWRVLGIMHLAETRDQAIADCTYG-LPDYAHYFGAVGVLPLANSADGADG
Mflv_2404|M.gilvum_PYR-GCK LLAIGGDLSMMVAKTDEEALKRLG-IGGGFFSFGIMHYYLTGMHTPGRTG
Mvan_4242|M.vanbaalenii_PYR-1 ILAIGGDLSMMVAKTDEEALERLG-IGGGFFSFGIMHYYLTGMHTPGRTG
MSMEG_4863|M.smegmatis_MC2_155 ILAIGGDLSMMVARTDEEALKRLG-IGGGFFSFGIMHYYLTGMHTPGRTK
TH_3770|M.thermoresistible__bu ILAIGGDLSMMVAKTDEEALKRLG-IGGGFFSFGIMHYYMTGMHTPGRTG
MMAR_4774|M.marinum_M ILAIGGDLSMMVAKTDEQALQRLG-QGGGFFSFGIMHYYMTGIHTPGRTG
MUL_0345|M.ulcerans_Agy99 ILAIGGDLSMMVAKTDEQALQRLG-QGGGFFSFGIMHYYMTGIHTPGRTG
MAV_0881|M.avium_104 ILAIGGDLSMMVAKTDEEALQRLG-KGGGFFSFGIMHYYMTGVHTPGRTG
Mb1971|M.bovis_AF2122/97 DWRLVREVLVAETDEQAFRYAVDGTMGRAMREYVLPTFRMFGMTKFYKHN
MLBr_00348|M.leprae_Br4923 DFEICASAQIVVTEDRAAAFAGIK--PFLALYMGGMGAEGTNFHADVYRR
: : :
Rv3618|M.tuberculosis_H37Rv ---SSREFVEQYAAKGNCCIGTPDDAIAHIEDLLHRSGGFG----TLLLL
MAB_0527c|M.abscessus_ATCC_199 A-IDPHEFVKAYADEGNCCIGTPEDAIEYIQGLLDTSGGFG----TFLML
Mflv_2404|M.gilvum_PYR-GCK ---VWELYEKAVKEDPTLAYGPGRGAIGSPETVREFLRGYEASGVDEIIL
Mvan_4242|M.vanbaalenii_PYR-1 ---VWELYEKAVKEDPTLAYGPGRGAIGSPDTVREFLRGYEASGVDEIIL
MSMEG_4863|M.smegmatis_MC2_155 ---VWERYEQAVADDPTLAYGPGRGAIGSPETVREFLRGYEASGVDEIIL
TH_3770|M.thermoresistible__bu ---VWELYEQAVKEDPTLAYGPGRGAIGSPETVREFLRGYEESGVDEIIL
MMAR_4774|M.marinum_M ---VWTRYLEEIEKDPTLAYGPGRGAIGCPATVREFLRGYEESGVDEIIL
MUL_0345|M.ulcerans_Agy99 ---VWTRYLEEIEKDPTLAYGPGRGAIGCPATVREFLRGYEESGVDEIIL
MAV_0881|M.avium_104 ---VWKRYLEEVEKDPTLAYGPGRGAIGSPATVREFLRGYEESGVDEIIL
Mb1971|M.bovis_AF2122/97 PSVPDDEVTPEYLAENTFVVGSVQTVVDKLEATYDQVGGFG----HLLIL
MLBr_00348|M.leprae_Br4923 -----MGYAEVVDEVTALFRSNQKDKAAEIIPNELVDSTMIVGDVDYVCK
. :
Rv3618|M.tuberculosis_H37Rv GHDWAPPPATFHS-YELFARAVIPYFKG--------QLAAPRASHEWARG
MAB_0527c|M.abscessus_ATCC_199 GHDWAAPEATFKS-YELFARKVIPHFTG--------QLTAAQASHDWAQA
Mflv_2404|M.gilvum_PYR-GCK LLNPRSHEGTMES-IEIMGRDILPEFIERDEKAVKDKAKRLEPVIEKVEA
Mvan_4242|M.vanbaalenii_PYR-1 LLNPRSHEGTMES-IEIMGRDILPEFIERDEKAVKDKAKRLEPVIEKVEA
MSMEG_4863|M.smegmatis_MC2_155 LLNPRSHEGTMES-IEIMGKEILPEFIERDEKAVKDKAERLEPVVEKVES
TH_3770|M.thermoresistible__bu LLNPRSHEGTMES-IEIMGREILPEFIEREEKAQVEKAKRLEPVLEKVEA
MMAR_4774|M.marinum_M LLNPRSHEGTMES-IELMGKEVLPEFIERDAKAVADKAVRLAPAIEKVQA
MUL_0345|M.ulcerans_Agy99 LLNPRSHEGTMES-IELMGKEVLPEFIERDAKAVADKAVRLAPAIEKVQA
MAV_0881|M.avium_104 LLNPRSHEGTMES-IELMGKEVLPEFIERDAKAVADKATRLAPVIEKVEA
Mb1971|M.bovis_AF2122/97 GFDYSDNPGPWKESLRLLAHEVMPRLNAR---------LATKPATAVV--
MLBr_00348|M.leprae_Br4923 QIAAWSAAGVTMMMVSASSVEQVRDLAGLF--------------------
. . : :
Rv3618|M.tuberculosis_H37Rv KR---------------DQLIGRAGEAVVKAITEHVAEQGEAGS------
MAB_0527c|M.abscessus_ATCC_199 KR---------------GELLGRAGDAVAKAIGEHVAQKASH--------
Mflv_2404|M.gilvum_PYR-GCK RRQSWDAPMFDETYSFGGLPTGRGGKFTAGEIPEAMAEINEGRVKAAQAE
Mvan_4242|M.vanbaalenii_PYR-1 RRQSWDAPLFDETYAFGGLPTGRGGKFTAGEIPEAMAEINEGRVKAAQAE
MSMEG_4863|M.smegmatis_MC2_155 RRQGSDAPLFDETYAFGGLPTGRGGKFTAGEIPEAMAEINEGRVKAAQAE
TH_3770|M.thermoresistible__bu RRKPSEAPEFDETYSFGGLPTGRGGKFTASEIPEAMAEINEGRVQAAKKA
MMAR_4774|M.marinum_M RRQESNAPQFDEAYSFGGLPTGRGGKFTASEIPEAMAEINEGRVQAAQRL
MUL_0345|M.ulcerans_Agy99 RRQESNAPQFDEAYSFGGLPTGRGGKFTASEIPEAMAEINEGRVQAAQRL
MAV_0881|M.avium_104 RRPKSTAPQFDENYSFGGLPTGRGGKFTASEIPEAMAEINEGRVQAAQRA
Mb1971|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00348|M.leprae_Br4923 --------------------------------------------------
Rv3618|M.tuberculosis_H37Rv --------------
MAB_0527c|M.abscessus_ATCC_199 --------------
Mflv_2404|M.gilvum_PYR-GCK KDARAAKEAAG---
Mvan_4242|M.vanbaalenii_PYR-1 KDAKAAKEAAS---
MSMEG_4863|M.smegmatis_MC2_155 K---AAREAAG---
TH_3770|M.thermoresistible__bu K------EQAS---
MMAR_4774|M.marinum_M K------EQRK---
MUL_0345|M.ulcerans_Agy99 K------EQRK---
MAV_0881|M.avium_104 K------EQQQQKK
Mb1971|M.bovis_AF2122/97 --------------
MLBr_00348|M.leprae_Br4923 --------------