For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NLGRNLSGPMQAVGALFSMSLDAIMFAFRRPFQGREFLDQCWFIARVSMAPTLLVAIPFTVLVSFTLNIL LRELGAADLSGAGAAFGAVTQLGPMVTVLIVAGAGATAMCADLGSRTIREEIDAMEVLGINPIQRLVTPR ILASGLVALLLNSLVVIIGILGGYVFSVFIQDVNPGAFAAGITLLTGVPEVIISCIKAALFGLIAGLVAC YRGLTIVGGGAKAVGNAVNETVVYAFMALFVINVVVTAIGIRMTTG*
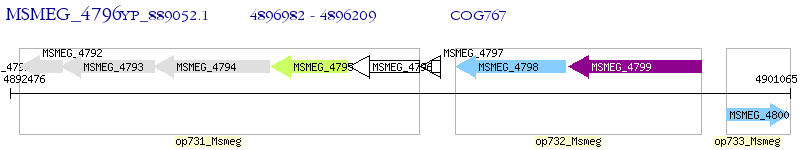
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4796 | - | - | 100% (257) | hypothetical protein MSMEG_4796 |
| M. smegmatis MC2 155 | MSMEG_2853 | - | e-124 | 90.40% (250) | hypothetical protein MSMEG_2853 |
| M. smegmatis MC2 155 | MSMEG_5902 | - | 1e-99 | 72.80% (250) | hypothetical protein MSMEG_5902 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3531c | yrbE4A | 1e-101 | 73.31% (251) | integral membrane protein YrbE4a |
| M. gilvum PYR-GCK | Mflv_2506 | - | 1e-127 | 89.49% (257) | hypothetical protein Mflv_2506 |
| M. tuberculosis H37Rv | Rv3501c | yrbE4A | 1e-101 | 73.31% (251) | integral membrane protein YrbE4a |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 4e-87 | 62.55% (251) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_4155c | - | 1e-100 | 73.81% (252) | hypothetical protein MAB_4155c |
| M. marinum M | MMAR_4712 | - | 1e-124 | 89.41% (255) | hypothetical protein MMAR_4712 |
| M. avium 104 | MAV_0946 | - | 1e-125 | 90.00% (250) | TrnB1 protein |
| M. thermoresistible (build 8) | TH_0523 | - | 1e-128 | 92.00% (250) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE4A |
| M. ulcerans Agy99 | MUL_4064 | yrbE4A | 1e-100 | 72.91% (251) | membrane protein YrbE4A |
| M. vanbaalenii PYR-1 | Mvan_4152 | - | 1e-127 | 89.88% (257) | hypothetical protein Mvan_4152 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3531c|M.bovis_AF2122/97 -------------------------MIQQ---LAVPARAVGGFFEMSMDT
MUL_4064|M.ulcerans_Agy99 MKASADPGLALGKHWRRRRLGRGTELIQQ---LAVPARAVGGFFEMSIDT
Rv3501c|M.tuberculosis_H37Rv -------------------------MIQQ---LAVPARAVGGFFEMSMDT
MAB_4155c|M.abscessus_ATCC_199 -------------------------MKQQ---LAAPARAVGGFVSMSLAT
MMAR_4712|M.marinum_M -----------MANRAADRWTPGITFGRG----SKPMQAVGGLFAMSADA
TH_0523|M.thermoresistible__bu -----------------------VVLSRG----SKPMQAIGGLFAMSLDA
MAV_0946|M.avium_104 -----------MADLRAERWSTGLPALRLKVDVSGAMQAVGALFAMSADA
Mflv_2506|M.gilvum_PYR-GCK -----------MAVKGPERWTTGISLPNG---LSGAMQAVGGFFSMALDA
Mvan_4152|M.vanbaalenii_PYR-1 -----------MATKGPERWTTGIALPNG---LSGAMAAVGGFFSMALDA
MSMEG_4796|M.smegmatis_MC2_155 -----------------------MNLGRN---LSGPMQAVGALFSMSLDA
MLBr_02587|M.leprae_Br4923 --------------MTTETRSSLVEYAHT--KLQTPLVLVGGFFRMLVLT
. . :*.:. * :
Mb3531c|M.bovis_AF2122/97 ARAAFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLRE
MUL_4064|M.ulcerans_Agy99 ARAAFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLRE
Rv3501c|M.tuberculosis_H37Rv ARAAFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLRE
MAB_4155c|M.abscessus_ATCC_199 FRNIFRRPFQGQEFLDQTWFIARVSLLPTLLVAIPFTVLVAFTLNILLRE
MMAR_4712|M.marinum_M IKFAFRRPFQWREFLEQSWFVARVAMLPTLLVAIPFTVLVSFTLNILLRE
TH_0523|M.thermoresistible__bu LKFVFRRPFQGREFLEQSWFIARVSLAPTLLVAIPFTVLVSFTLNILLRE
MAV_0946|M.avium_104 VKYLFRRPFQWREFLEQSWFVARVSLAPTLLVAIPFTVLVSFTLNILLRE
Mflv_2506|M.gilvum_PYR-GCK VRFVFVRPFQWREFLEQSWFVAKVSMAPTLLVAIPFTVLVSFTLNILLRE
Mvan_4152|M.vanbaalenii_PYR-1 VRFVFVRPFQWREFLEQCWFVARVSMAPTLLVAIPFTVLVSFTLNILLRE
MSMEG_4796|M.smegmatis_MC2_155 IMFAFRRPFQGREFLDQCWFIARVSMAPTLLVAIPFTVLVSFTLNILLRE
MLBr_02587|M.leprae_Br4923 GKALFRRPFQWREFILQCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQ
* **** :**: * *:: :*:: **::*:**:***: ****:** :
Mb3531c|M.bovis_AF2122/97 IGAADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEID
MUL_4064|M.ulcerans_Agy99 IGAADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEID
Rv3501c|M.tuberculosis_H37Rv IGAADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEID
MAB_4155c|M.abscessus_ATCC_199 IGAADLSGAATAFGTVTQLGPVVTVLVVAGAGATAICADLGARTIREEID
MMAR_4712|M.marinum_M LGAADLSGAGAAFGAVTQVGPLVTVLIVAGAGATAMCADLGSRTIREEID
TH_0523|M.thermoresistible__bu LGAADLSGAGAAFGAVTQVGPMVTVLIVAGAGATAMCADLGSRTIREEID
MAV_0946|M.avium_104 LGAADLSGAGAAFGAVTQLGPLVTVLIVAGAGATAMCADLGSRTIREEID
Mflv_2506|M.gilvum_PYR-GCK LGAADLSGAGAAFGAVTQVGPLVTVLIVAGAGATAMCADLGSRTIREEID
Mvan_4152|M.vanbaalenii_PYR-1 LGAADLSGAGAAFGAVTQVGPLVTVLIVAGAGATAMCADLGSRTIREEID
MSMEG_4796|M.smegmatis_MC2_155 LGAADLSGAGAAFGAVTQLGPMVTVLIVAGAGATAMCADLGSRTIREEID
MLBr_02587|M.leprae_Br4923 FSAADLSGAGAAIGAVTQLGPLTTVLVVAGAGSTSICADLGARTIREEID
:.*******.:*:*::**:**:.***:*****:*::*****:********
Mb3531c|M.bovis_AF2122/97 AMRVLGIDPIQRLVVPRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGV
MUL_4064|M.ulcerans_Agy99 AMRVLGIDPIQRLVVPRVLASTVVALLLNGLVCAIGLSGGYAFSVFLQGV
Rv3501c|M.tuberculosis_H37Rv AMRVLGIDPIQRLVVPRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGV
MAB_4155c|M.abscessus_ATCC_199 AMQVLGIDPIQRLVVPRVLASTFVALLLNGLVCAIGMVGGYVFSVFLQGV
MMAR_4712|M.marinum_M AMEVLGINPVQRLVTPRMLASGLVALLLNSLVVIIGIVGGYVFSVFVQDV
TH_0523|M.thermoresistible__bu AMEVLGINPVQRLVTPRMLASGLVALLLNSLVVIIGIIGGYTFSVFVQDV
MAV_0946|M.avium_104 AMEVLGINPIQRLVTPRMLASGLVAFLLNSLVVIIGVLGGYVFSVFVQDV
Mflv_2506|M.gilvum_PYR-GCK AMEVLGINPVQRLVTPRMLASGLVALLLNSLVVIIGILGGYFFSIFVQDV
Mvan_4152|M.vanbaalenii_PYR-1 AMEVLGINPVQRLVTPRMLAAGLVALLLNSLVVIIGILGGYFFSVFVQDV
MSMEG_4796|M.smegmatis_MC2_155 AMEVLGINPIQRLVTPRILASGLVALLLNSLVVIIGILGGYVFSVFIQDV
MLBr_02587|M.leprae_Br4923 AMEVLGIDPIHRLVVPRVLAATLVATLLNGLVITVGLVGGYLFGVYLQNV
**.****:*::***.**:**: .** ***.** :*: *** *.:::*.*
Mb3531c|M.bovis_AF2122/97 NPGAFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGG-PKG
MUL_4064|M.ulcerans_Agy99 NPGAFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGG-PKG
Rv3501c|M.tuberculosis_H37Rv NPGAFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGG-PKG
MAB_4155c|M.abscessus_ATCC_199 NPGAFINGLTVLTGLGELMISEVKAFLFGIFAGLVGCYRGLTVKGG-PKG
MMAR_4712|M.marinum_M NPGAFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLTISGGGAKA
TH_0523|M.thermoresistible__bu NPGAFAAGITLLTGVPEVVISCVKAALFGLIAGLVACYRGLTISGGGAKA
MAV_0946|M.avium_104 NPGAFAAGITLLTGVPEVVISCIKAALFGLIAGLVACYRGLSITGGGAKA
Mflv_2506|M.gilvum_PYR-GCK NPGAFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLTITGGGAKA
Mvan_4152|M.vanbaalenii_PYR-1 NPGAFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLTITGGGAKA
MSMEG_4796|M.smegmatis_MC2_155 NPGAFAAGITLLTGVPEVIISCIKAALFGLIAGLVACYRGLTIVGGGAKA
MLBr_02587|M.leprae_Br4923 SGGAYLATLTTITGLPEVVIATVKAATFGLIAGLVGCYRGLIVRGG-SNG
. **: :* :**: *:::: :** **::****.***** : ** .:.
Mb3531c|M.bovis_AF2122/97 VGNAVNETVVYAFICLFVINVVMTAIGVRISAQ-
MUL_4064|M.ulcerans_Agy99 VGNAVNETVVYAFICLFVINVVMTAIGVRISAR-
Rv3501c|M.tuberculosis_H37Rv VGNAVNETVVYAFICLFVINVVMTAIGVRISAQ-
MAB_4155c|M.abscessus_ATCC_199 VGDAVNETVVYAFICLFVINVVMTAIGVRVLVKH
MMAR_4712|M.marinum_M VGNAVNETVVYAFMSLFVVNVVVTAIGIRMSAK-
TH_0523|M.thermoresistible__bu VGNAVNETVVYAFMALFVINVVVTAIGIRLTTG-
MAV_0946|M.avium_104 VGNAVNETVVYAFMSLFVVNVVVTAIGIKMTAK-
Mflv_2506|M.gilvum_PYR-GCK VGNAVNETVVYAFMALFVINVVVTAIGIRMTTG-
Mvan_4152|M.vanbaalenii_PYR-1 VGNAVNETVVYAFMALFVINVVVTAIGIRMTTG-
MSMEG_4796|M.smegmatis_MC2_155 VGNAVNETVVYAFMALFVINVVVTAIGIRMTTG-
MLBr_02587|M.leprae_Br4923 LGAAVNETVVLCVVALYAVNVVLTTIGVRFGTGH
:* ******* ..:.*:.:***:*:**::. .