For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADLRAERWSTGLPALRLKVDVSGAMQAVGALFAMSADAVKYLFRRPFQWREFLEQSWFVARVSLAPTLL VAIPFTVLVSFTLNILLRELGAADLSGAGAAFGAVTQLGPLVTVLIVAGAGATAMCADLGSRTIREEIDA MEVLGINPIQRLVTPRMLASGLVAFLLNSLVVIIGVLGGYVFSVFVQDVNPGAFAAGITLLTGVPEVVIS CIKAALFGLIAGLVACYRGLSITGGGAKAVGNAVNETVVYAFMSLFVVNVVVTAIGIKMTAK
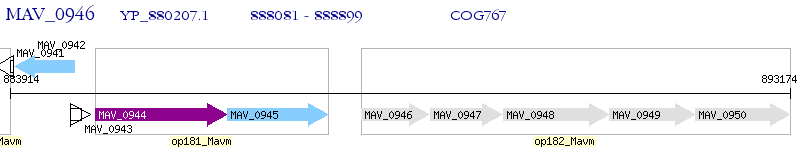
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0946 | - | - | 100% (272) | TrnB1 protein |
| M. avium 104 | MAV_0655 | - | 5e-96 | 70.33% (246) | hypothetical protein MAV_0655 |
| M. avium 104 | MAV_2059 | - | 1e-88 | 60.96% (251) | TrnB1 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3531c | yrbE4A | 3e-98 | 71.95% (246) | integral membrane protein YrbE4a |
| M. gilvum PYR-GCK | Mflv_2506 | - | 1e-128 | 86.30% (270) | hypothetical protein Mflv_2506 |
| M. tuberculosis H37Rv | Rv3501c | yrbE4A | 3e-98 | 71.95% (246) | integral membrane protein YrbE4a |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 2e-85 | 64.17% (240) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_4155c | - | 6e-97 | 69.41% (255) | hypothetical protein MAB_4155c |
| M. marinum M | MMAR_4712 | - | 1e-129 | 87.13% (272) | hypothetical protein MMAR_4712 |
| M. smegmatis MC2 155 | MSMEG_4796 | - | 1e-125 | 90.00% (250) | hypothetical protein MSMEG_4796 |
| M. thermoresistible (build 8) | TH_0393 | - | 1e-126 | 84.50% (271) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE4A |
| M. ulcerans Agy99 | MUL_4064 | yrbE4A | 6e-98 | 71.54% (246) | membrane protein YrbE4A |
| M. vanbaalenii PYR-1 | Mvan_2849 | - | 1e-128 | 86.35% (271) | hypothetical protein Mvan_2849 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3531c|M.bovis_AF2122/97 -------------------------MIQQLAVPARAVGGFFEMSMDTARA
MUL_4064|M.ulcerans_Agy99 MKASADPGLALGKHWRRRRLGRGTELIQQLAVPARAVGGFFEMSIDTARA
Rv3501c|M.tuberculosis_H37Rv -------------------------MIQQLAVPARAVGGFFEMSMDTARA
MAB_4155c|M.abscessus_ATCC_199 -------------------------MKQQLAAPARAVGGFVSMSLATFRN
MAV_0946|M.avium_104 --------MADLRAERWSTGLPALRLKVDVSGAMQAVGALFAMSADAVKY
MMAR_4712|M.marinum_M --------MANRAADRWTPGITFGRG----SKPMQAVGGLFAMSADAIKF
TH_0393|M.thermoresistible__bu --------MVNKAVDRWSTGLPLPAGG--TAGTLRAVGGLFSMSVDAVRF
Mvan_2849|M.vanbaalenii_PYR-1 --------MVDKAADRWSSGLPALGAG--MAAPMQGVGGLLSMSADAVKF
Mflv_2506|M.gilvum_PYR-GCK --------MAVKGPERWTTGISLPNG---LSGAMQAVGGFFSMALDAVRF
MSMEG_4796|M.smegmatis_MC2_155 ---------MNLGRN--------------LSGPMQAVGALFSMSLDAIMF
MLBr_02587|M.leprae_Br4923 -----------MTTETRSSLVEYAHTK--LQTPLVLVGGFFRMLVLTGKA
. **.:. * :
Mb3531c|M.bovis_AF2122/97 AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
MUL_4064|M.ulcerans_Agy99 AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
Rv3501c|M.tuberculosis_H37Rv AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
MAB_4155c|M.abscessus_ATCC_199 IFRRPFQGQEFLDQTWFIARVSLLPTLLVAIPFTVLVAFTLNILLREIGA
MAV_0946|M.avium_104 LFRRPFQWREFLEQSWFVARVSLAPTLLVAIPFTVLVSFTLNILLRELGA
MMAR_4712|M.marinum_M AFRRPFQWREFLEQSWFVARVAMLPTLLVAIPFTVLVSFTLNILLRELGA
TH_0393|M.thermoresistible__bu AFRRPFQGREFLEQSWFVARVALLPTLLVAIPFTVLVSFTLNILLRELGA
Mvan_2849|M.vanbaalenii_PYR-1 LFRRPFQGREFLEQSWFVARVSLMPTLLVAIPFTVLVSFTLNILLRELGA
Mflv_2506|M.gilvum_PYR-GCK VFVRPFQWREFLEQSWFVAKVSMAPTLLVAIPFTVLVSFTLNILLRELGA
MSMEG_4796|M.smegmatis_MC2_155 AFRRPFQGREFLDQCWFIARVSMAPTLLVAIPFTVLVSFTLNILLRELGA
MLBr_02587|M.leprae_Br4923 LFRRPFQWREFILQCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSA
* **** :**: * *:: :*:: **::*:**:***: ****:** ::.*
Mb3531c|M.bovis_AF2122/97 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MUL_4064|M.ulcerans_Agy99 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
Rv3501c|M.tuberculosis_H37Rv ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MAB_4155c|M.abscessus_ATCC_199 ADLSGAATAFGTVTQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMQ
MAV_0946|M.avium_104 ADLSGAGAAFGAVTQLGPLVTVLIVAGAGATAMCADLGSRTIREEIDAME
MMAR_4712|M.marinum_M ADLSGAGAAFGAVTQVGPLVTVLIVAGAGATAMCADLGSRTIREEIDAME
TH_0393|M.thermoresistible__bu ADLSGAGAAFGAVTQVGPMVTVLIVAGAGATAMCADLGSRTIRDEVDALE
Mvan_2849|M.vanbaalenii_PYR-1 ADLSGAGAAFGAVTQVGPMVTVLIVAGAGATAMCADLGSRTIREEIDALE
Mflv_2506|M.gilvum_PYR-GCK ADLSGAGAAFGAVTQVGPLVTVLIVAGAGATAMCADLGSRTIREEIDAME
MSMEG_4796|M.smegmatis_MC2_155 ADLSGAGAAFGAVTQLGPMVTVLIVAGAGATAMCADLGSRTIREEIDAME
MLBr_02587|M.leprae_Br4923 ADLSGAGAAIGAVTQLGPLTTVLVVAGAGSTSICADLGARTIREEIDAME
******.:*:*::**:**:.***:*****:*::*****:****:*:**:.
Mb3531c|M.bovis_AF2122/97 VLGIDPIQRLVVPRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
MUL_4064|M.ulcerans_Agy99 VLGIDPIQRLVVPRVLASTVVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
Rv3501c|M.tuberculosis_H37Rv VLGIDPIQRLVVPRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
MAB_4155c|M.abscessus_ATCC_199 VLGIDPIQRLVVPRVLASTFVALLLNGLVCAIGMVGGYVFSVFLQGVNPG
MAV_0946|M.avium_104 VLGINPIQRLVTPRMLASGLVAFLLNSLVVIIGVLGGYVFSVFVQDVNPG
MMAR_4712|M.marinum_M VLGINPVQRLVTPRMLASGLVALLLNSLVVIIGIVGGYVFSVFVQDVNPG
TH_0393|M.thermoresistible__bu VLGINPVQRLVTPRMLASGLVALLLNSLVVIIGILGGYTFSVFVQDVNPG
Mvan_2849|M.vanbaalenii_PYR-1 VLGINPVQRLVTPRMLASGLVALLLNSLVVIIGILGGYFFSIFIQGVNPG
Mflv_2506|M.gilvum_PYR-GCK VLGINPVQRLVTPRMLASGLVALLLNSLVVIIGILGGYFFSIFVQDVNPG
MSMEG_4796|M.smegmatis_MC2_155 VLGINPIQRLVTPRILASGLVALLLNSLVVIIGILGGYVFSVFIQDVNPG
MLBr_02587|M.leprae_Br4923 VLGIDPIHRLVVPRVLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGG
****:*::***.**:**: .** ***.** :*: *** *.:::*.*. *
Mb3531c|M.bovis_AF2122/97 AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGG-PKGVGN
MUL_4064|M.ulcerans_Agy99 AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGG-PKGVGN
Rv3501c|M.tuberculosis_H37Rv AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGG-PKGVGN
MAB_4155c|M.abscessus_ATCC_199 AFINGLTVLTGLGELMISEVKAFLFGIFAGLVGCYRGLTVKGG-PKGVGD
MAV_0946|M.avium_104 AFAAGITLLTGVPEVVISCIKAALFGLIAGLVACYRGLSITGGGAKAVGN
MMAR_4712|M.marinum_M AFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLTISGGGAKAVGN
TH_0393|M.thermoresistible__bu AFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLTISGGGAKAVGN
Mvan_2849|M.vanbaalenii_PYR-1 AFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLSISGGGAKAVGN
Mflv_2506|M.gilvum_PYR-GCK AFAAGITLLTGVPEVIISCVKAALFGLIAGLVACYRGLTITGGGAKAVGN
MSMEG_4796|M.smegmatis_MC2_155 AFAAGITLLTGVPEVIISCIKAALFGLIAGLVACYRGLTIVGGGAKAVGN
MLBr_02587|M.leprae_Br4923 AYLATLTTITGLPEVVIATVKAATFGLIAGLVGCYRGLIVRGG-SNGLGA
*: :* :**: *:::: :** **::****.***** : ** .:.:*
Mb3531c|M.bovis_AF2122/97 AVNETVVYAFICLFVINVVMTAIGVRISAQ-
MUL_4064|M.ulcerans_Agy99 AVNETVVYAFICLFVINVVMTAIGVRISAR-
Rv3501c|M.tuberculosis_H37Rv AVNETVVYAFICLFVINVVMTAIGVRISAQ-
MAB_4155c|M.abscessus_ATCC_199 AVNETVVYAFICLFVINVVMTAIGVRVLVKH
MAV_0946|M.avium_104 AVNETVVYAFMSLFVVNVVVTAIGIKMTAK-
MMAR_4712|M.marinum_M AVNETVVYAFMSLFVVNVVVTAIGIRMSAK-
TH_0393|M.thermoresistible__bu AVNETVVYAFMALFVVNVVVTAIGIRMTAG-
Mvan_2849|M.vanbaalenii_PYR-1 AVNETVVYAFMALFVVNVVVTAIGIRMTSG-
Mflv_2506|M.gilvum_PYR-GCK AVNETVVYAFMALFVINVVVTAIGIRMTTG-
MSMEG_4796|M.smegmatis_MC2_155 AVNETVVYAFMALFVINVVVTAIGIRMTTG-
MLBr_02587|M.leprae_Br4923 AVNETVVLCVVALYAVNVVLTTIGVRFGTGH
******* ..:.*:.:***:*:**::.