For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KASADPGLALGKHWRRRRLGRGTELIQQLAVPARAVGGFFEMSIDTARAAFRRPFQFREFLDQTWMVARV SLVPTLLVSIPFTVLVAFTLNILLREIGAADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGART IREEIDAMRVLGIDPIQRLVVPRVLASTVVALLLNGLVCAIGLSGGYAFSVFLQGVNPGAFINGLTVLTG LRELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNAVNETVVYAFICLFVINVVMTAIGVRISAR*
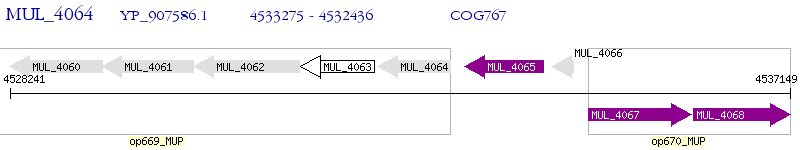
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4064 | yrbE4A | - | 100% (279) | membrane protein YrbE4A |
| M. ulcerans Agy99 | MUL_1060 | yrbE1A | 5e-90 | 64.00% (250) | integral membrane protein YrbE1A |
| M. ulcerans Agy99 | MUL_3916 | yrbE3A_1 | 5e-87 | 60.48% (248) | integral membrane protein YrbE3A_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3531c | yrbE4A | 1e-137 | 98.82% (254) | integral membrane protein YrbE4a |
| M. gilvum PYR-GCK | Mflv_1554 | - | 1e-120 | 92.41% (237) | hypothetical protein Mflv_1554 |
| M. tuberculosis H37Rv | Rv3501c | yrbE4A | 1e-137 | 98.82% (254) | integral membrane protein YrbE4a |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 1e-89 | 64.37% (247) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_4155c | - | 1e-121 | 87.25% (251) | hypothetical protein MAB_4155c |
| M. marinum M | MMAR_4989 | yrbE4A | 1e-128 | 100.00% (237) | membrane protein YrbE4A |
| M. avium 104 | MAV_0655 | - | 1e-134 | 95.67% (254) | hypothetical protein MAV_0655 |
| M. smegmatis MC2 155 | MSMEG_5902 | - | 1e-128 | 91.34% (254) | hypothetical protein MSMEG_5902 |
| M. thermoresistible (build 8) | TH_1939 | - | 1e-125 | 90.84% (251) | CONSERVED HYPOTHETICAL INTEGRAL MEMBRANE PROTEIN YRBE4A |
| M. vanbaalenii PYR-1 | Mvan_5204 | - | 1e-120 | 92.74% (234) | hypothetical protein Mvan_5204 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1554|M.gilvum_PYR-GCK ------------------------------------------MSLDTFVK
Mvan_5204|M.vanbaalenii_PYR-1 ------------------------------------------MSLDTFVK
MSMEG_5902|M.smegmatis_MC2_155 -------------------------MIEQLAVPARAVGGFVEMSLDTFAK
Mb3531c|M.bovis_AF2122/97 -------------------------MIQQLAVPARAVGGFFEMSMDTARA
Rv3501c|M.tuberculosis_H37Rv -------------------------MIQQLAVPARAVGGFFEMSMDTARA
MUL_4064|M.ulcerans_Agy99 MKASADPGLALGKHWRRRRLGRGTELIQQLAVPARAVGGFFEMSIDTARA
MMAR_4989|M.marinum_M ------------------------------------------MSIDTARA
MAV_0655|M.avium_104 -------------------------MIEQLAVPARAVGGFFEMMIDTGRA
TH_1939|M.thermoresistible__bu -------------------------LIQQLAVPARAVGGFMEMSGETFVK
MAB_4155c|M.abscessus_ATCC_199 -------------------------MKQQLAAPARAVGGFVSMSLATFRN
MLBr_02587|M.leprae_Br4923 -------------MTTETRSSLVEYAHTKLQTPLVLVGGFFRMLVLTGKA
* *
Mflv_1554|M.gilvum_PYR-GCK TFRRPFQFREFLEQTWMIARVSLVPTLLVAIPFTVLVAFTLNILLREIGA
Mvan_5204|M.vanbaalenii_PYR-1 TFRRPFQFREFLEQTWMIARVSLVPTLLVAIPFTVLVAFTLNILLREIGA
MSMEG_5902|M.smegmatis_MC2_155 IFRRPFQLKEFLDQTWMIARVSLVPTLLVAIPFTVLVAFTLNILLREIGA
Mb3531c|M.bovis_AF2122/97 AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
Rv3501c|M.tuberculosis_H37Rv AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
MUL_4064|M.ulcerans_Agy99 AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
MMAR_4989|M.marinum_M AFRRPFQFREFLDQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
MAV_0655|M.avium_104 AFRRPFQFGEFLDQTWMIARVSLVPTLLVSIPFTVLVAFTLNILLREIGA
TH_1939|M.thermoresistible__bu IFRRPFQLREFLDQTWMIARVSLIPTLLVSIPFTVLVAFTLNILLREIGA
MAB_4155c|M.abscessus_ATCC_199 IFRRPFQGQEFLDQTWFIARVSLLPTLLVAIPFTVLVAFTLNILLREIGA
MLBr_02587|M.leprae_Br4923 LFRRPFQWREFILQCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSA
****** **: * *:: **:::**::*:**:***: ****:** ::.*
Mflv_1554|M.gilvum_PYR-GCK ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
Mvan_5204|M.vanbaalenii_PYR-1 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MSMEG_5902|M.smegmatis_MC2_155 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
Mb3531c|M.bovis_AF2122/97 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
Rv3501c|M.tuberculosis_H37Rv ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MUL_4064|M.ulcerans_Agy99 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MMAR_4989|M.marinum_M ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MAV_0655|M.avium_104 ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
TH_1939|M.thermoresistible__bu ADLSGAGTAFGTITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMR
MAB_4155c|M.abscessus_ATCC_199 ADLSGAATAFGTVTQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMQ
MLBr_02587|M.leprae_Br4923 ADLSGAGAAIGAVTQLGPLTTVLVVAGAGSTSICADLGARTIREEIDAME
******.:*:*::*****:.*********:*:*****************.
Mflv_1554|M.gilvum_PYR-GCK VLGIDPIQRLVVPRVLASTFVALLLNGLVCLIGLSGGYVFSVFLQGVNPG
Mvan_5204|M.vanbaalenii_PYR-1 VLGIDPIQRLVVPRVLASTFVALLLNGLVCLIGLSGGYVFSVFLQGVNPG
MSMEG_5902|M.smegmatis_MC2_155 VLGIDPIQRLVVPRVLASTFVALLLNGLVCAIGLAGGYVFSVFLQGVNPG
Mb3531c|M.bovis_AF2122/97 VLGIDPIQRLVVPRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
Rv3501c|M.tuberculosis_H37Rv VLGIDPIQRLVVPRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
MUL_4064|M.ulcerans_Agy99 VLGIDPIQRLVVPRVLASTVVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
MMAR_4989|M.marinum_M VLGIDPIQRLVVPRVLASTVVALLLNGLVCAIGLSGGYAFSVFLQGVNPG
MAV_0655|M.avium_104 VLGIDPIHRLVVPRVLASTVVALLLNGLVCAIGLSGGYVFSVFLQGVNPG
TH_1939|M.thermoresistible__bu VLGIDPIQRLVVPRVLASTFVALLLNGLVCAIGLSGGYVFSVLLQGVNPG
MAB_4155c|M.abscessus_ATCC_199 VLGIDPIQRLVVPRVLASTFVALLLNGLVCAIGMVGGYVFSVFLQGVNPG
MLBr_02587|M.leprae_Br4923 VLGIDPIHRLVVPRVLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGG
*******:*********:*.** ****** :*: *** *.* **.*. *
Mflv_1554|M.gilvum_PYR-GCK SFINGLTVLTGLPELILAEIKALLFGVVAGLVGCYRGLTVKGGPKGVGNA
Mvan_5204|M.vanbaalenii_PYR-1 AFINGLTVLTGLGELVLAEIKALLFGVVAGLVGCYRGLTVKGGPKGVGNA
MSMEG_5902|M.smegmatis_MC2_155 AFINGLTVLTGLGELVLAEIKALLFGVVAGLVGCYRGLTVKGGPKGVGNA
Mb3531c|M.bovis_AF2122/97 AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNA
Rv3501c|M.tuberculosis_H37Rv AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNA
MUL_4064|M.ulcerans_Agy99 AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNA
MMAR_4989|M.marinum_M AFINGLTVLTGLRELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNA
MAV_0655|M.avium_104 AFINGLTVLTGLRELVLAEVKALLFGVMAGMVGCYRGLTVKGGPKGVGNA
TH_1939|M.thermoresistible__bu AFVNGLTVLTGLPELLLAEVKALLFGVTAGLVGCYRGLTVKGGPKGVGEA
MAB_4155c|M.abscessus_ATCC_199 AFINGLTVLTGLGELMISEVKAFLFGIFAGLVGCYRGLTVKGGPKGVGDA
MLBr_02587|M.leprae_Br4923 AYLATLTTITGLPEVVIATVKAATFGLIAGLVGCYRGLIVRGGSNGLGAA
::: **.:*** *:::: :** **: **:******* *:**.:*:* *
Mflv_1554|M.gilvum_PYR-GCK VNETVVYAFICLFVINVIMTAIGVRVLVR-
Mvan_5204|M.vanbaalenii_PYR-1 VNETVVYAFICLFVINVIMTAVGVRVLDS-
MSMEG_5902|M.smegmatis_MC2_155 VNETVVYAFICLFVINVIMTAVGVRVLTR-
Mb3531c|M.bovis_AF2122/97 VNETVVYAFICLFVINVVMTAIGVRISAQ-
Rv3501c|M.tuberculosis_H37Rv VNETVVYAFICLFVINVVMTAIGVRISAQ-
MUL_4064|M.ulcerans_Agy99 VNETVVYAFICLFVINVVMTAIGVRISAR-
MMAR_4989|M.marinum_M VNETVVYAFICLFVINVVMTAIGVRISAR-
MAV_0655|M.avium_104 VNETVVYAFICLFVINVVMTAIGVRISAK-
TH_1939|M.thermoresistible__bu VNETVVYAFICLFVINVVMTALGTQVLD--
MAB_4155c|M.abscessus_ATCC_199 VNETVVYAFICLFVINVVMTAIGVRVLVKH
MLBr_02587|M.leprae_Br4923 VNETVVLCVVALYAVNVVLTTIGVRFGTGH
****** ..:.*:.:**::*::*.:.