For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDSTATDPAATTPDFDTVDYFTDQSLVPDPHPYFDHLRSKCPVVREPHYGVLAITSFEEATTVLKDTET FSSCIAVGGPFPPLPFTPEGDDITGQIEQHRTQLPMFEHMVTMDPPEHTNARSLLNRLLTPKRLKENEDF MWRLADECLDDFIDDGSCEFLKQYAKPFSLLVIADLLGVPEEDHDEFRHVLGAPRPGAIVGSLDGDQLAM NPLAWLDDKFVRYLEDRRKEPRDDVLTALATAKYPDGSTPEVIDVVRSATFLFAAGQETTTKLLSASLRV LGDRPDIQQALREDRSRIPTFVEEALRMDAPVKSQFRLAKKTTQLGGVDVPAGTTLMVCPGAVNRDPVRF EDPHTFSLDRKNVREHIAFGRGVHSCPGGPLARVEGRVSLERILDRMADIRIDEEHHGPADNRRYTYEPT YILRGLTDLHIKFEPVR
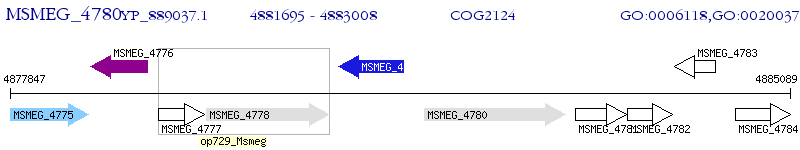
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4780 | - | - | 100% (437) | cytochrome p450 |
| M. smegmatis MC2 155 | MSMEG_6030 | - | e-158 | 61.27% (426) | cytochrome p450 |
| M. smegmatis MC2 155 | MSMEG_4823 | - | e-139 | 56.51% (430) | cytochrome p450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1806 | cyp144 | 4e-46 | 32.05% (390) | cytochrome p450 144 CYP144 |
| M. gilvum PYR-GCK | Mflv_2517 | - | 0.0 | 81.97% (427) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1777 | cyp144 | 2e-45 | 31.79% (390) | cytochrome p450 144 CYP144 |
| M. leprae Br4923 | MLBr_02088 | - | 3e-33 | 29.00% (331) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1216c | - | 7e-40 | 28.43% (415) | cytochrome P450 |
| M. marinum M | MMAR_4694 | cyp150A6 | 0.0 | 72.14% (420) | cytochrome P450 150A6 Cyp150A6 |
| M. avium 104 | MAV_0964 | - | 0.0 | 73.27% (419) | cytochrome p450 |
| M. thermoresistible (build 8) | TH_4542 | - | 0.0 | 73.82% (424) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0314 | cyp150A6 | 0.0 | 70.95% (420) | cytochrome P450 150A6 Cyp150A6 |
| M. vanbaalenii PYR-1 | Mvan_4141 | - | 0.0 | 83.84% (427) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1806|M.bovis_AF2122/97 -MRRSPKGSPGAVLDLQRRVDQAVSADHAELMTIAKDANTFFGAESVQDP
Rv1777|M.tuberculosis_H37Rv -MRRSPKGSPGAVLDLQRRVDQAVSADHAELMTIAKDANTFFGAESVQDP
Mflv_2517|M.gilvum_PYR-GCK -------------------------------MTDFDTIDYFTDPSLVPDP
Mvan_4141|M.vanbaalenii_PYR-1 -------------------------------MSDFDTIDYFTDPSLVPDP
MSMEG_4780|M.smegmatis_MC2_155 -------------------MTDSTATDPAATTPDFDTVDYFTDQSLVPDP
TH_4542|M.thermoresistible__bu ------------------------------MTTDFDSIDYFTDPSLVPDP
MMAR_4694|M.marinum_M -------------------------------MSSYESIDFFTDPSLIPDP
MUL_0314|M.ulcerans_Agy99 -------------------------------MSSYESIDFFTDPSLIPDP
MAV_0964|M.avium_104 -------------------------------MSNFDSIDFFTDPSLVPDP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
MAB_1216c|M.abscessus_ATCC_199 --------------------------MTVAETSAVPLVFDPYDYDFHEDP
. **
Mb1806|M.bovis_AF2122/97 YPLYERMRAAGSVHRIANSDFYAVCGWDAVNEAIGRPEDFSSNLTAT---
Rv1777|M.tuberculosis_H37Rv YPLYERMRAAGSVHRIANSDFYAVCGWDAVNEAIGRPEDFSSNLTAT---
Mflv_2517|M.gilvum_PYR-GCK HPYFDHLRSKCPVVKEPHYGVLAVTGYEEAATVLKDSDTFSSCIAVAGPF
Mvan_4141|M.vanbaalenii_PYR-1 HPYFDHLRSKCPVVREPHYGVLAVTGYEEAATVLKDSDTFSSCIAVAGPF
MSMEG_4780|M.smegmatis_MC2_155 HPYFDHLRSKCPVVREPHYGVLAITSFEEATTVLKDTETFSSCIAVGGPF
TH_4542|M.thermoresistible__bu HPYFDHMRSKCPIAREPHHGVIAVTGWEEANDVYRDSENLSSCIAVMGPF
MMAR_4694|M.marinum_M HPYFDYLRSQSPVLRLPQYGVVAVTGYEEATAVYKDTDSFSNCVALGGPF
MUL_0314|M.ulcerans_Agy99 HPYFDYLRRQSPVLRLPQYGVVAVTGYEEATAVYKDTDSFSNCVALGGPF
MAV_0964|M.avium_104 HPYFDYLRSQNPVLRLPHYGVVAITGYEEATEVYKDPETFSNIVALGGPF
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPG-MPLTVFSSFSDCDEALRHPLSASDRLKAT---
MAB_1216c|M.abscessus_ATCC_199 YPYYRRLRDEAPLYRNDDLKFWALSRHHDVLQGFRNSEALSNANGVS---
.* : : .: . .. . *.
Mb1806|M.bovis_AF2122/97 --MTYTAEGTAKPFEMDPLGGPT---HVLATADDPAHAVHRKLVLRHLAA
Rv1777|M.tuberculosis_H37Rv --MTYTAEGTAKPFEMDPLGGPT---HVLATADDPAHAVHRKLVLRHLAA
Mflv_2517|M.gilvum_PYR-GCK PPLPFTPEGDDITDQITAHRPQMPMFEHMVTMDPPDHTNARSLLNRLLTP
Mvan_4141|M.vanbaalenii_PYR-1 PPLPFTPEGDDITGQIAAHRSQMPMFEHMVTMDPPDHTNARSLLNRLLTP
MSMEG_4780|M.smegmatis_MC2_155 PPLPFTPEGDDITGQIEQHRTQLPMFEHMVTMDPPEHTNARSLLNRLLTP
TH_4542|M.thermoresistible__bu TPLPFQPEGDDITEQIARHRTQIPMYEHMVTMDPPEHTNARSILARLLTP
MMAR_4694|M.marinum_M PPLPFAPNGDDVNAQIDAHREQFPMYEHMVTMDPPEHSRARSILSRLLTP
MUL_0314|M.ulcerans_Agy99 PPLPFTPNGDDVNAQIDAHREQFPMYEHMVTMDPPEHSRARSILSRLLTP
MAV_0964|M.avium_104 PPLPFTPEGDDISAQIDAHRSSFPMFEHMVTMDPPEHTKARSVLAKLLTP
MLBr_02088|M.leprae_Br4923 ---------LAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAP
MAB_1216c|M.abscessus_ATCC_199 ---------MDKASFGPHAK----LVMSFLAMDDPEHLRLRALVSKGFTP
: * * * * :: : ::.
Mb1806|M.bovis_AF2122/97 KRIRVMEQFTVQAADRLWVDGMQD---GCIEWMGAMANRLPMMVVAELIG
Rv1777|M.tuberculosis_H37Rv KRIRVMEQFTVQAADRLWVDGMQD---GCIEWMGAMANRLPMMVVAELIG
Mflv_2517|M.gilvum_PYR-GCK SRLKENEDFMWRLADEVLDDIIGAEPAGRCEFLTAYAKPFSLLVIADMLG
Mvan_4141|M.vanbaalenii_PYR-1 SRLKENEDFMWRLADEVLDDIISRDPVGRCEFLSAYAKPFSLLVIADMLG
MSMEG_4780|M.smegmatis_MC2_155 KRLKENEDFMWRLADECLDDFIDD---GSCEFLKQYAKPFSLLVIADLLG
TH_4542|M.thermoresistible__bu KRLKENEDFMWRLADRHIDEFIAD---GKCEFLAAYAKPFSLLVVADLLG
MMAR_4694|M.marinum_M SRLKQNEEFMWRLADRQLDEFLGA---GECEFISEYAKPFATLVIADLLG
MUL_0314|M.ulcerans_Agy99 SRLKQNEEFMWRLADRQLDEFLGA---AECEFISEYAKPFATLVIADLLG
MAV_0964|M.avium_104 SRLKQNEEFMWRLADRQLDEFLHN---GECEFIAEYSKPFATLVIADLLG
MLBr_02088|M.leprae_Br4923 KVVQALEGDIAALVDSLLDKGAAAG---QFDVIADLAFPLAVAVICRLLG
MAB_1216c|M.abscessus_ATCC_199 RRIRELEGQVVALARTHLDRALANASDRSFDFIAEYAGKLPMDVISELMG
:: * . : : : :. *:. ::*
Mb1806|M.bovis_AF2122/97 LPDPDIAQLVKWGYAATQLLEGLVENDQ-----LVAAGVALMELSGYIFE
Rv1777|M.tuberculosis_H37Rv LPDPDIAQLVKWGYAATQLLEGLVENDQ-----LVAAGVALMELSGYIFE
Mflv_2517|M.gilvum_PYR-GCK VPEADHEEFRTVLGAPRPGANVGSLDHS-----DLVGANPLEWLDEKFIG
Mvan_4141|M.vanbaalenii_PYR-1 VPEADHEEFRTVLGAPRPGANVGSLDHS-----DLVGANPLEWLDEKFIG
MSMEG_4780|M.smegmatis_MC2_155 VPEEDHDEFRHVLGAPRPGAIVGSLDG------DQLAMNPLAWLDDKFVR
TH_4542|M.thermoresistible__bu VPEEDHDAFREAFGAERPGTRIGSLDH------ETIATNPLEWADDKFTQ
MMAR_4694|M.marinum_M VPEDDRKDFRVVLGADRMG-RVGALDH------ESVGVNPLQWLDDKFSA
MUL_0314|M.ulcerans_Agy99 VPEDDRKDFRVVLGADRMG-RVGALDH------ESVGVNPLQWLDDKFSA
MAV_0964|M.avium_104 VPEEDHTQFRTVLGADRPGARVGALDH------ETVGINPLEWLDDKFCN
MLBr_02088|M.leprae_Br4923 VPYEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQ
MAB_1216c|M.abscessus_ATCC_199 VPESDRARIRELADGVMHREDGLADVP-------PEAIQASFDLMTYYIE
:* * : .
Mb1806|M.bovis_AF2122/97 QFDRAAADPRDNLLGELATACASG--ELDTLTAQVMMVTLFAAGGESTAA
Rv1777|M.tuberculosis_H37Rv QFDRAAADPRDNLLGELATACASG--ELDTLTAQVMMVTLFAAGGESTAA
Mflv_2517|M.gilvum_PYR-GCK YLEDRRKEPRDDVLTALATAKYPDGSTPPVIEVVRSATFLFAAGQETTTK
Mvan_4141|M.vanbaalenii_PYR-1 YLEDRRKEPRDDVLTALATAKYPDGSTPPVIEVVRSATFLFAAGQETTTK
MSMEG_4780|M.smegmatis_MC2_155 YLEDRRKEPRDDVLTALATAKYPDGSTPEVIDVVRSATFLFAAGQETTTK
TH_4542|M.thermoresistible__bu YIEDRRREPRDDVLTAIATAKYPDGSTPEVIDVVRTATFLFAAGQETTAK
MMAR_4694|M.marinum_M YIEDRRRQPRNDVLTALATATYPDGSTPEVIDVVRSATFLFAAGQETTAK
MUL_0314|M.ulcerans_Agy99 YIEDRRRRPRNDVLTALATATYPDGSTPEIIDVVRWATFLFAAGQETTAK
MAV_0964|M.avium_104 YIEERRREPRADVLTFLAEAKYPDGSTPPVIEVVRSATFLFAAGQETTAK
MLBr_02088|M.leprae_Br4923 LVKCRRGTPGEDLISRLIELDESG-DQLTEEEIIATCGLLLVAGHETTVN
MAB_1216c|M.abscessus_ATCC_199 MVRERRRRPTEDLTSALLQAEIDG-DRLTDEEVLAFLFLMVIAGNETTTK
. * :: : . :. ** *:*.
Mb1806|M.bovis_AF2122/97 LLGSAVWILATRPDIQQQVRANPELLGAFIEETLRYEPPFRGQYRHVRNA
Rv1777|M.tuberculosis_H37Rv LLGSAVWILATRPDIQQQVRANPELLGAFIEETLRYEPPFRGHYRHVRNA
Mflv_2517|M.gilvum_PYR-GCK LLSASMRVLGDHPEIQERLRRDRTRIPIFIEEALRMDAPVKSQFRLAKKN
Mvan_4141|M.vanbaalenii_PYR-1 LLSASLRVLGDHPDIQERLRRDRSRIPIFVEEALRMDAPVKSQFRLAKKN
MSMEG_4780|M.smegmatis_MC2_155 LLSASLRVLGDRPDIQQALREDRSRIPTFVEEALRMDAPVKSQFRLAKKT
TH_4542|M.thermoresistible__bu LLGAALQILGDRPDIQQTLREDRSRIPVFIEECLRMESPVKTVFRMVRKS
MMAR_4694|M.marinum_M LLTAAMRVLGDRPDIQRRLRENRSLIPNFIEESLRMDSPVKSDSRLARKR
MUL_0314|M.ulcerans_Agy99 QLTAAMRVLGDRPDIQRQLRENRSLIPNFIEESLRMDSPVKSDSRLARKR
MAV_0964|M.avium_104 LLSAALQVLGDRPDIQQQLREDRSLIPAFIEESLRMESPVKSDSRLARRA
MLBr_02088|M.leprae_Br4923 LIANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKD
MAB_1216c|M.abscessus_ATCC_199 LLANAVYWGHRNPDQLATVHADHDRIPLWVEESLRYDTSSQILARTVAED
: :: .*. : : :** ** :.. : * . .
Mb1806|M.bovis_AF2122/97 TTLDGTELPADSHLLLLWGAANRDPAQFEAPGEFRLDRAGGKGHISFGKG
Rv1777|M.tuberculosis_H37Rv TTLDGTELPADSHLLLLWGAANRDPAQFEAPGEFRLDRAGGKGHISFGKG
Mflv_2517|M.gilvum_PYR-GCK TKVGDMDVPAGTTMMVCPGAVNRDPSRFENPHDFDLDRKNVREHIAFGRG
Mvan_4141|M.vanbaalenii_PYR-1 TRVGDMDVPAGTTMMVCPGAVNRDPSRFEHPHEFDLDRKNVREHIAFGRG
MSMEG_4780|M.smegmatis_MC2_155 TQLGGVDVPAGTTLMVCPGAVNRDPVRFEDPHTFSLDRKNVREHIAFGRG
TH_4542|M.thermoresistible__bu TTVGDLDAPAGTTVMVAPGAANRDPRRFENPHEFDIDRKNVREHLAFSRG
MMAR_4694|M.marinum_M TTVGGLDIAAGTVVMVLPGAANRDPRRFEDPHEFRLDRPNVREHMAFARG
MUL_0314|M.ulcerans_Agy99 TTVGGLDIAAGTVVMVLSGAANRDPRRFEDPHEFRLDRPNVREHMAFARG
MAV_0964|M.avium_104 TTIGGVDIPAGTVVMILPGAANRDPRRFENPHEFDLRRKNVREHMAFARG
MLBr_02088|M.leprae_Br4923 MTIGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPS-SRHLAFAVG
MAB_1216c|M.abscessus_ATCC_199 ITVYDTKIPAGDILLLLPGSANRDDRVFDDADQYRIGREIGAKLVSFGSG
: . . ::: .:.*** :. . : * ::*. *
Mb1806|M.bovis_AF2122/97 AHFCVGAALARLEARIVLRLLLDRTSVIEAADVGG---------WLPSIL
Rv1777|M.tuberculosis_H37Rv AHFCVGAALARLEARIVLRLLLDRTSVIEAADVGG---------WLPSIL
Mflv_2517|M.gilvum_PYR-GCK VHSCPGGPLARVEGRVSIERILDRMGDIAIDEELHGPPGDRRYNYEPTFI
Mvan_4141|M.vanbaalenii_PYR-1 VHSCPGGPLARVEGRVSIERILDRMGDIAIDEELHGPPGARRYNYEPTFI
MSMEG_4780|M.smegmatis_MC2_155 VHSCPGGPLARVEGRVSLERILDRMADIRIDEEHHGPADNRRYTYEPTYI
TH_4542|M.thermoresistible__bu IHSCPGAPLARVEGRVSIERLLDRLADIEIDADKHGGPGERNYTYEPTFI
MMAR_4694|M.marinum_M VHSCPGGPLARVEGRVSLERILDRMLDIAINEDRHGPADDRRYTYEPTYI
MUL_0314|M.ulcerans_Agy99 VHSCPGGPLARVEGRVSLERILDRMLDIAINEDRHGPADDRRYTYEPTYI
MAV_0964|M.avium_104 VHSCPGGPLARVEGRVSIERILDRMPHIEINETHHGPAGDRRYTYEPTYI
MLBr_02088|M.leprae_Br4923 SHFCLGAALARLEATVTLSAISARFPQVQLAG---------ELVYKPNVA
MAB_1216c|M.abscessus_ATCC_199 AHFCLGAHLARMEAKVALTELFTRISGYEIDER--------NSVRVHSSN
* * *. ***:*. : : : * .
Mb1806|M.bovis_AF2122/97 VRRIERLELAVQ---------
Rv1777|M.tuberculosis_H37Rv VRRIERLELAVQ---------
Mflv_2517|M.gilvum_PYR-GCK LRGLTDINITFTPIR------
Mvan_4141|M.vanbaalenii_PYR-1 LRGLTDLNITFTPVR------
MSMEG_4780|M.smegmatis_MC2_155 LRGLTDLHIKFEPVR------
TH_4542|M.thermoresistible__bu LRGLTELNITFKPIG------
MMAR_4694|M.marinum_M LRGLTELHITFTPAG------
MUL_0314|M.ulcerans_Agy99 LRGLTELHITFTPAG------
MAV_0964|M.avium_104 LRGLSELHLTFTTADAVAPVG
MLBr_02088|M.leprae_Br4923 MRGMSALPVQV----------
MAB_1216c|M.abscessus_ATCC_199 VRGFAHLPMTVQLREGR----
:* : : : .