For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPSGRVQKTAGTPHGSGATAVGSADAAPRIELVGVRKVFGDGRREVVAVDHADLTVADGELFAILGPSGS GKTTVLRMIAGFETPTSGTIRLGGTDVTGVPARHRDVNTVFQEYALFPHMTVAQNIEYGLRVRGVGKAER RRRAADALDMVRLTDQARRRPIQLSGGQRQRVALARALVGRPRVLLLDEPLGALDLKLREQMQVELKAIQ REVGITFVIVTHDQDEALTLCDRLAVFNNGRIEQVGGAREVYEHPANRFVADFVGTSNVLDGEDAVAVFG RSGTLAVRPERIAVAPAGTEPAAGHRGIAATVTEIIYAGPLTRIAATTAGGVAVNASVLTSSSSLPQDIS HGSPITLTWPESAVHHLD
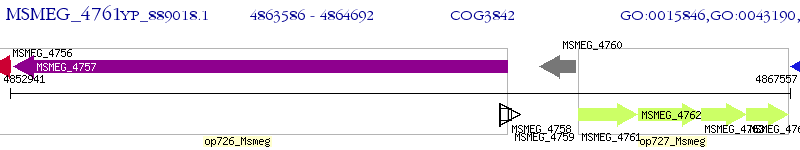
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4761 | - | - | 100% (368) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_3281 | - | 1e-85 | 47.23% (379) | spermidine/putrescine ABC transporter ATP-binding subunit |
| M. smegmatis MC2 155 | MSMEG_0662 | - | 2e-78 | 48.63% (329) | putrescine transport ATP-binding protein PotG |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 9e-65 | 45.11% (317) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_3138 | - | 1e-85 | 48.14% (376) | spermidine/putrescine ABC transporter ATPase subunit |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 1e-64 | 45.11% (317) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01424 | - | 3e-57 | 45.39% (271) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_1655 | - | 2e-68 | 44.14% (333) | sulfate ABC transporter, ATP-binding protein SysA |
| M. marinum M | MMAR_3715 | cysA1 | 1e-65 | 45.35% (333) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 3e-64 | 45.00% (320) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. thermoresistible (build 8) | TH_2424 | - | 8e-64 | 42.90% (331) | PUTATIVE sulfate ABC transporter, ATPase subunit |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 6e-65 | 45.43% (328) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_3387 | - | 8e-86 | 50.97% (361) | spermidine/putrescine ABC transporter ATPase subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3138|M.gilvum_PYR-GCK MRNRLRRTAESAIVYPQPGGSIEEDGALTRAHTATGHGIGQGAASTKGGP
Mvan_3387|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4761|M.smegmatis_MC2_155 ----------------MPSGRVQKTAGTPHGSGATAVG------SADAAP
Mb2419c|M.bovis_AF2122/97 ---------------------------------MT--------------Y
Rv2397c|M.tuberculosis_H37Rv ---------------------------------MT--------------Y
MMAR_3715|M.marinum_M ---------------------------MKKETSLTGPGT--------GGH
MUL_3658|M.ulcerans_Agy99 ---------------------------------MTGPGT--------GGH
MAV_1785|M.avium_104 ---------------------------------MTDAGTD------RGDI
TH_2424|M.thermoresistible__bu ---------------------------------MT--------------D
MAB_1655|M.abscessus_ATCC_1997 ---------------------------------MT------------AEN
MLBr_01424|M.leprae_Br4923 ------------------------------------------------MA
Mflv_3138|M.gilvum_PYR-GCK VIEIDHVTKRFGD----YLAVSDADFSIASGEFFSMLGPSGCGKTTTLRM
Mvan_3387|M.vanbaalenii_PYR-1 MIEIDHVTKSFGD----YVAVADADFSIASGEFFSMLGPSGCGKTTTLRM
MSMEG_4761|M.smegmatis_MC2_155 RIELVGVRKVFGDGRREVVAVDHADLTVADGELFAILGPSGSGKTTVLRM
Mb2419c|M.bovis_AF2122/97 AIVVADATKRYGD----FVALDHVDFVVPTGSLTALLGPSGSGKSTLLRT
Rv2397c|M.tuberculosis_H37Rv AIVVADATKRYGD----FVALDHVDFVVPTGSLTALLGPSGSGKSTLLRT
MMAR_3715|M.marinum_M AIVVRDAFKQYGD----FVALDHVDFVVPAGSLTALLGPSGSGKSTLLRT
MUL_3658|M.ulcerans_Agy99 AIVVRDAFKQYGD----FVALDHVDFVVPAGSLTALLGPSGSGKSTLLRT
MAV_1785|M.avium_104 AITVRDAYKRYGD----FVALDHVDFVVPTGSLTALLGPSGSGKSTLLRT
TH_2424|M.thermoresistible__bu AITVRGANKRYGD----FAALDNVDFAVPAGSLTALLGPSGSGKSTLLRA
MAB_1655|M.abscessus_ATCC_1997 TITVRGANKHYGD----FAALDNIDFDVPSGSLTALLGPSGSGKSTLLRS
MLBr_01424|M.leprae_Br4923 SVTFEQATRQFPDT--DRPALDRLDLDVDDGEFVVVVGPSGCGKTTSLRM
: . . : : * *: *: : *.: ::****.**:* **
Mflv_3138|M.gilvum_PYR-GCK IAGFETPSEGAIRLEGADVSKTPPNKRNVNTVFQHYALFPHMTVWDNVAY
Mvan_3387|M.vanbaalenii_PYR-1 IAGFETPTEGAIRLEGADVSRTPPNKRNVNTVFQHYALFPHMTVWDNVAY
MSMEG_4761|M.smegmatis_MC2_155 IAGFETPTSGTIRLGGTDVTGVPARHRDVNTVFQEYALFPHMTVAQNIEY
Mb2419c|M.bovis_AF2122/97 IAGLDQPDTGTITINGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAF
Rv2397c|M.tuberculosis_H37Rv IAGLDQPDTGTITINGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAF
MMAR_3715|M.marinum_M IAGLDQPDSGNITINGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAY
MUL_3658|M.ulcerans_Agy99 IAGLDQPDSGNITINGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAY
MAV_1785|M.avium_104 IAGLDQPDTGTVTIYGRDVTRVPPQRRGIGFVFQHYAAFKHLTVRDNVAY
TH_2424|M.thermoresistible__bu IAGLDRPDTGTITINGRDVTDVPPQRRGIGFVFQHYAAFKHLTVRDNVAF
MAB_1655|M.abscessus_ATCC_1997 IAGLDNPDSGTITINGRDVTGVPPQRRDIGFVFQHYAAFKHLTVWENVAF
MLBr_01424|M.leprae_Br4923 VAGLETLDSGCIRIGDRDVTHVDPKDRDVAMVFQNYALYPHMTVAQNLGF
:**:: * : : . **: . .. *.: ***.** : *:** :*: :
Mflv_3138|M.gilvum_PYR-GCK GPRSKKLDKGEIRKRVDELLEIVRLSDFAERKPAQLSGGQQQRVALARAL
Mvan_3387|M.vanbaalenii_PYR-1 GPRSKKLGKGEIRKRVDELLDIVRLTDFAERKPAQLSGGQQQRVALARAL
MSMEG_4761|M.smegmatis_MC2_155 GLRVRGVGKAERRRRAADALDMVRLTDQARRRPIQLSGGQRQRVALARAL
Mb2419c|M.bovis_AF2122/97 GLKIRKRPKAEIKAKVDNLLQVVGLSGFQSRYPNQLSGGQRQRMALARAL
Rv2397c|M.tuberculosis_H37Rv GLKIRKRPKAEIKAKVDNLLQVVGLSGFQSRYPNQLSGGQRQRMALARAL
MMAR_3715|M.marinum_M GLKIRKRPKAEIRDKVDNLLEVVGLSGFHGRYPNQLSGGQRQRMALARAL
MUL_3658|M.ulcerans_Agy99 GLKIRKRPKAEIRDKVDNLLEVVGLSGFHGRYPDQLSGGQRQRMALARAL
MAV_1785|M.avium_104 GLKVRKRPKAEIKAKVDNLLEVVGLSGFQGRYPNQLSGGQRQRMALARAL
TH_2424|M.thermoresistible__bu GLKIRRRPKREIAEKVDNLLEVVGLAGFQNRYPNQLSGGQRQRMALARAL
MAB_1655|M.abscessus_ATCC_1997 GLKIRKKPKAEIKKKVDDLLEVVGLSGFHTRYPAQLSGGQRQRMALARAL
MLBr_01424|M.leprae_Br4923 ALKISKISKAEIHDRILVVAKLLDLQPYLDRKPKDLSGGQRQRVAMGRAI
. : * * : .:: * * * :*****:**:*:.**:
Mflv_3138|M.gilvum_PYR-GCK VNYPSALLLDEPLGALDLKLRHVMQFELKRIQREIGITFIYVTHDQEEAL
Mvan_3387|M.vanbaalenii_PYR-1 VNYPSALLLDEPLGALDLKLRHVMQFELKRIQREVGITFIYVTHDQEEAL
MSMEG_4761|M.smegmatis_MC2_155 VGRPRVLLLDEPLGALDLKLREQMQVELKAIQREVGITFVIVTHDQDEAL
Mb2419c|M.bovis_AF2122/97 AVDPEVLLLDEPFGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEAL
Rv2397c|M.tuberculosis_H37Rv AVDPEVLLLDEPFGALDAKVREELRAWLRRLHDEVHVTTVLVTHDQAEAL
MMAR_3715|M.marinum_M AVDPEVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEAL
MUL_3658|M.ulcerans_Agy99 AVDPEVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEAL
MAV_1785|M.avium_104 AVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEAL
TH_2424|M.thermoresistible__bu AVDPEVLLLDEPFGALDAKVREDLRTWLRRLHEEVHVTTVLVTHDQSEAL
MAB_1655|M.abscessus_ATCC_1997 AVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEAL
MLBr_01424|M.leprae_Br4923 VRRPQVFLMDEPLSNLDAKLRAQTRNQIAELQCQLGTTTVYVTHDQVEAM
. * .:*:***:. ** *:* : : :: :: * : ***** **:
Mflv_3138|M.gilvum_PYR-GCK TMSDRIAVMNAGNVEQIGTPTEIYDRPATVFVASFIGQANLWAGRQTGRT
Mvan_3387|M.vanbaalenii_PYR-1 TMSDRIAVMNAGNVEQIGTPTEIYDRPATVFVASFIGQANLWAGRQTGRA
MSMEG_4761|M.smegmatis_MC2_155 TLCDRLAVFNNGRIEQVGGAREVYEHPANRFVADFVGTSNVLDGED----
Mb2419c|M.bovis_AF2122/97 DVADRIAVLHKGRIEQVGSPTDVYDAPANAFVMSFLGAVSTLNGSLVRP-
Rv2397c|M.tuberculosis_H37Rv DVADRIAVLHKGRIEQVGSPTDVYDAPANAFVMSFLGAVSTLNGSLVRP-
MMAR_3715|M.marinum_M DVADRIAVLNSGRIEQVGSPTEVYDAPANAFVMSFLGAVSALNGALVRP-
MUL_3658|M.ulcerans_Agy99 DVADRIAVLNSGRIEQVGSPTEVYDAPTNAFVMSFLGAVSALNGALVRP-
MAV_1785|M.avium_104 DVADRIAVLNQGRIEQIGSPTEVYDAPTNAFVMSFLGAVSTLNGTLVRP-
TH_2424|M.thermoresistible__bu DVADRIAVLNNGRIEQVGSPAEVYDSPANEFVMSFLGAVSSLNGTLVRP-
MAB_1655|M.abscessus_ATCC_1997 DVADRIAVLNKGRIEQVGSPEDLYDRPANSFVMSFLGPVAKLNGILVRP-
MLBr_01424|M.leprae_Br4923 TMGDRVAVLCYGVLQQFATPRELYHKPENVFVAGFIGSPGMNLVTLSIVD
: **:**: * ::*.. . ::*. * . ** .*:*
Mflv_3138|M.gilvum_PYR-GCK NRDFVEIDVLGSTLKARPGETTIEPGGHATLMVRPERVRVS--MDAPTGD
Mvan_3387|M.vanbaalenii_PYR-1 NRDYVEVDVLGSTLKARPGETTIEPGGHATLMVRPERIRVS--MDAPTGD
MSMEG_4761|M.smegmatis_MC2_155 -----AVAVFG-----RSG----------TLAVRPERIAVAPAGTEPAAG
Mb2419c|M.bovis_AF2122/97 ---------------------------HDIRVGRTPNMAVAA-ADGTAGS
Rv2397c|M.tuberculosis_H37Rv ---------------------------HDIRVGRTPNMAVAA-ADGTAGS
MMAR_3715|M.marinum_M ---------------------------HDIRVGRTPEMAVAA-ADGTGES
MUL_3658|M.ulcerans_Agy99 ---------------------------HDIRVGRTPEMAVAA-ADGTGES
MAV_1785|M.avium_104 ---------------------------HDIRVGRTPEMAVAA-EDGTAES
TH_2424|M.thermoresistible__bu ---------------------------HDIWVGRHPELPTS---DSTLQA
MAB_1655|M.abscessus_ATCC_1997 ---------------------------HDIRVGRNADLARAA-AEGTGET
MLBr_01424|M.leprae_Br4923 ---------------------------SSVLLGDWPIRIPREIASAASEV
.
Mflv_3138|M.gilvum_PYR-GCK VAGVRATVSDLTFQGPVVRLSLAAPD-DSTVIAHVGP-EQDLPLLRPGDE
Mvan_3387|M.vanbaalenii_PYR-1 VAAVRATVSDLTFQGPVVRLSLAAPD-DSTVIAHVGP-EQDLPLLRPGDD
MSMEG_4761|M.smegmatis_MC2_155 HRGIAATVTEIIYAGPLTRIAATTAG-GVAVNASVLT-SSSS---LPQD-
Mb2419c|M.bovis_AF2122/97 TGVLRAVVDRVVVLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDT
Rv2397c|M.tuberculosis_H37Rv TGVLRAVVDRVVVLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDT
MMAR_3715|M.marinum_M TGVTRAVVDRIVVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDT
MUL_3658|M.ulcerans_Agy99 TGVTRAVVDRIVVLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDT
MAV_1785|M.avium_104 TGVARAIVDRVVKLGFEVRVELTSAATGGPFTAQITRGDAEALALREGDT
TH_2424|M.thermoresistible__bu TGVVRARIDRVVMLGFEVRLELTSATNNAPFTAQITRGDAEALGLRSGDT
MAB_1655|M.abscessus_ATCC_1997 TGVTKATVDRVVYLGFEVRVELTSAATGEQFTAQITRGDAEALGLREGDP
MLBr_01424|M.leprae_Br4923 IIGVRPEHFELGSLGVEVEIDMVEELGADAYLYGRIAGASKVTDQLVVAR
. : * ..: . .
Mflv_3138|M.gilvum_PYR-GCK VYVSWAPDASLVLPGADIPTTEDLEEMLDDS---------
Mvan_3387|M.vanbaalenii_PYR-1 VFVSWAPDASLVLPGADIPTTEDLEEMLDDS---------
MSMEG_4761|M.smegmatis_MC2_155 --ISHGSPITLTWP-------ESAVHHLD-----------
Mb2419c|M.bovis_AF2122/97 VYVRATRVPPIAG---GVSGVDDAGVERVKVTST------
Rv2397c|M.tuberculosis_H37Rv VYVRATRVPPIAG---GVSGVDDAGVERVKVTST------
MMAR_3715|M.marinum_M VYVRATRVPPIAG---AAPAVPDDGAGSTPSAAQIGVGGQ
MUL_3658|M.ulcerans_Agy99 VYVRATRVPPIAG---AAPAVPDDGAGSTPRAAQIGVGGQ
MAV_1785|M.avium_104 VYVRATRVPPITAGATTVPAVSRDGADEATLTSA------
TH_2424|M.thermoresistible__bu VYVRATRVPALPT-----GDVDERVAAR------------
MAB_1655|M.abscessus_ATCC_1997 VYVRVTRVPDVGE----LAEVAS-----------------
MLBr_01424|M.leprae_Br4923 VDGRNPPAKGSRVRLYTEPGNVHFFGVDGHRIS-------