For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAENTITVRGANKHYGDFAALDNIDFDVPSGSLTALLGPSGSGKSTLLRSIAGLDNPDSGTITINGRDV TGVPPQRRDIGFVFQHYAAFKHLTVWENVAFGLKIRKKPKAEIKKKVDDLLEVVGLSGFHTRYPAQLSGG QRQRMALARALAVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVL NKGRIEQVGSPEDLYDRPANSFVMSFLGPVAKLNGILVRPHDIRVGRNADLARAAAEGTGETTGVTKATV DRVVYLGFEVRVELTSAATGEQFTAQITRGDAEALGLREGDPVYVRVTRVPDVGELAEVAS
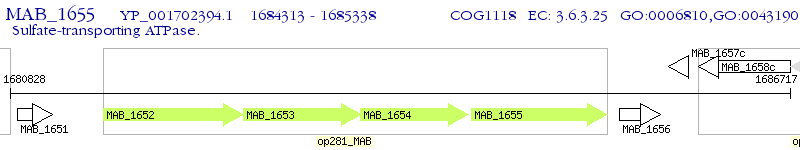
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1655 | - | - | 100% (341) | sulfate ABC transporter, ATP-binding protein SysA |
| M. abscessus ATCC 19977 | MAB_1502 | - | 9e-55 | 50.23% (219) | ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1375 | - | 4e-54 | 45.06% (233) | sugar ABC transporter, ATP-binding protein SugC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 1e-152 | 83.18% (327) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_2681 | - | 1e-152 | 79.82% (332) | sulfate ABC transporter, ATPase subunit |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 1e-152 | 83.18% (327) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01424 | - | 1e-54 | 43.01% (272) | putative binding-protein-dependent transport protein |
| M. marinum M | MMAR_3715 | cysA1 | 1e-155 | 82.14% (336) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 1e-152 | 82.57% (327) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. smegmatis MC2 155 | MSMEG_4530 | cysA | 1e-158 | 81.95% (338) | sulfate ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_2424 | - | 1e-147 | 80.12% (327) | PUTATIVE sulfate ABC transporter, ATPase subunit |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-154 | 81.85% (336) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_3872 | - | 1e-155 | 82.98% (329) | sulfate ABC transporter, ATPase subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2681|M.gilvum_PYR-GCK ----------MSATSELAIEVKGANKRFG--DFAALDDVDFTVPKGSLTS
Mvan_3872|M.vanbaalenii_PYR-1 --------------MTHAITVEGANKRYG--DFAALDNVDFVVPAGSLTA
TH_2424|M.thermoresistible__bu --------------MTDAITVRGANKRYG--DFAALDNVDFAVPAGSLTA
MSMEG_4530|M.smegmatis_MC2_155 ------MSDTSTQTNTSAIVVRGANKHYG--DFAALDNIDFEVPSGSLTA
Mb2419c|M.bovis_AF2122/97 ------MT--------YAIVVADATKRYG--DFVALDHVDFVVPTGSLTA
Rv2397c|M.tuberculosis_H37Rv ------MT--------YAIVVADATKRYG--DFVALDHVDFVVPTGSLTA
MMAR_3715|M.marinum_M MKKETSLTGPGT--GGHAIVVRDAFKQYG--DFVALDHVDFVVPAGSLTA
MUL_3658|M.ulcerans_Agy99 ------MTGPGT--GGHAIVVRDAFKQYG--DFVALDHVDFVVPAGSLTA
MAV_1785|M.avium_104 ------MTDAGTDRGDIAITVRDAYKRYG--DFVALDHVDFVVPTGSLTA
MAB_1655|M.abscessus_ATCC_1997 ------------MTAENTITVRGANKHYG--DFAALDNIDFDVPSGSLTA
MLBr_01424|M.leprae_Br4923 ---------------MASVTFEQATRQFPDTDRPALDRLDLDVDDGEFVV
:: . * ::: * *** :*: * *.:.
Mflv_2681|M.gilvum_PYR-GCK LLGPSGSGKSTLLRAIAGLDDPDTGTVIINGRDVTNVPPQRRGIGFVFQH
Mvan_3872|M.vanbaalenii_PYR-1 LLGPSGSGKSTLLRAIAGLDEPDTGTITINGRDVTRIPPQKRGIGFVFQH
TH_2424|M.thermoresistible__bu LLGPSGSGKSTLLRAIAGLDRPDTGTITINGRDVTDVPPQRRGIGFVFQH
MSMEG_4530|M.smegmatis_MC2_155 LLGPSGSGKSTLLRAIAGLDQPDSGTVTINGRDVTGVPPQQRGIGFVFQH
Mb2419c|M.bovis_AF2122/97 LLGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQRRGIGFVFQH
Rv2397c|M.tuberculosis_H37Rv LLGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQRRGIGFVFQH
MMAR_3715|M.marinum_M LLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQRRGIGFVFQH
MUL_3658|M.ulcerans_Agy99 LLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQRRGIGFVFQH
MAV_1785|M.avium_104 LLGPSGSGKSTLLRTIAGLDQPDTGTVTIYGRDVTRVPPQRRGIGFVFQH
MAB_1655|M.abscessus_ATCC_1997 LLGPSGSGKSTLLRSIAGLDNPDSGTITINGRDVTGVPPQRRDIGFVFQH
MLBr_01424|M.leprae_Br4923 VVGPSGCGKTTSLRMVAGLETLDSGCIRIGDRDVTHVDPKDRDVAMVFQN
::****.**:* ** :***: *:* : * .**** : *: *.:.:***:
Mflv_2681|M.gilvum_PYR-GCK YAAFKHLTVRDNVAFGLKIRKKPKAEIKEKVDNLLEVVGLAGFQGRYPNQ
Mvan_3872|M.vanbaalenii_PYR-1 YAAFKHLTVRDNVAFGLKIRKKPKAEIKAKVDDLLEVVGLAGFQSRYPNQ
TH_2424|M.thermoresistible__bu YAAFKHLTVRDNVAFGLKIRRRPKREIAEKVDNLLEVVGLAGFQNRYPNQ
MSMEG_4530|M.smegmatis_MC2_155 YAAFKHLSVRDNVAFGLKIRKRPKAEIAERVDNLLEVVGLSGFQTRYPNQ
Mb2419c|M.bovis_AF2122/97 YAAFKHLTVRDNVAFGLKIRKRPKAEIKAKVDNLLQVVGLSGFQSRYPNQ
Rv2397c|M.tuberculosis_H37Rv YAAFKHLTVRDNVAFGLKIRKRPKAEIKAKVDNLLQVVGLSGFQSRYPNQ
MMAR_3715|M.marinum_M YAAFKHLTVRDNVAYGLKIRKRPKAEIRDKVDNLLEVVGLSGFHGRYPNQ
MUL_3658|M.ulcerans_Agy99 YAAFKHLTVRDNVAYGLKIRKRPKAEIRDKVDNLLEVVGLSGFHGRYPDQ
MAV_1785|M.avium_104 YAAFKHLTVRDNVAYGLKVRKRPKAEIKAKVDNLLEVVGLSGFQGRYPNQ
MAB_1655|M.abscessus_ATCC_1997 YAAFKHLTVWENVAFGLKIRKKPKAEIKKKVDDLLEVVGLSGFHTRYPAQ
MLBr_01424|M.leprae_Br4923 YALYPHMTVAQNLGFALKISKISKAEIHDRILVVAKLLDLQPYLDRKPKD
** : *::* :*:.:.**: : .* ** :: : :::.* : * * :
Mflv_2681|M.gilvum_PYR-GCK LSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHEEV
Mvan_3872|M.vanbaalenii_PYR-1 LSGGQRQRMALARALAVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEV
TH_2424|M.thermoresistible__bu LSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRTWLRRLHEEV
MSMEG_4530|M.smegmatis_MC2_155 LSGGQRQRMALARALAVDPQVLLLDEPFGALDAKVRDDLRTWLRRLHDEV
Mb2419c|M.bovis_AF2122/97 LSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLHDEV
Rv2397c|M.tuberculosis_H37Rv LSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLHDEV
MMAR_3715|M.marinum_M LSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHDEV
MUL_3658|M.ulcerans_Agy99 LSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHDEV
MAV_1785|M.avium_104 LSGGQRQRMALARALAVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEV
MAB_1655|M.abscessus_ATCC_1997 LSGGQRQRMALARALAVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEV
MLBr_01424|M.leprae_Br4923 LSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRAQTRNQIAELQCQL
********:*:.**:. *:*:*:***:. ****:* : * : .*: ::
Mflv_2681|M.gilvum_PYR-GCK HVTTVLVTHDQSEALDISDRIAVLNKGRIEQVGSPSEVYDAPANAFVMSF
Mvan_3872|M.vanbaalenii_PYR-1 HVTTVLVTHDQQEALDVADRIAVLNKGRIEQVGSPTEVYDAPANAFVMSF
TH_2424|M.thermoresistible__bu HVTTVLVTHDQSEALDVADRIAVLNNGRIEQVGSPAEVYDSPANEFVMSF
MSMEG_4530|M.smegmatis_MC2_155 HVTTVLVTHDQSEALDVADRIAVLNKGRIEQIGSPTEVYDQPANAFVMSF
Mb2419c|M.bovis_AF2122/97 HVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFVMSF
Rv2397c|M.tuberculosis_H37Rv HVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFVMSF
MMAR_3715|M.marinum_M HVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPANAFVMSF
MUL_3658|M.ulcerans_Agy99 HVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPTNAFVMSF
MAV_1785|M.avium_104 HVTTVLVTHDQAEALDVADRIAVLNQGRIEQIGSPTEVYDAPTNAFVMSF
MAB_1655|M.abscessus_ATCC_1997 HVTTVLVTHDQAEALDVADRIAVLNKGRIEQVGSPEDLYDRPANSFVMSF
MLBr_01424|M.leprae_Br4923 GTTTVYVTHDQVEAMTMGDRVAVLCYGVLQQFATPRELYHKPENVFVAGF
.*** ***** **: :.**:*** * ::*..:* ::*. * * ** .*
Mflv_2681|M.gilvum_PYR-GCK LG--AVSSLNGVLVRPHDIRVGRNPDMAIATTDNSIQSTGVTRAVIERIV
Mvan_3872|M.vanbaalenii_PYR-1 LG--AVSSLNGVLVRPHDIRVGRSPEMAIGASDDSVQATGVTRATIDRIV
TH_2424|M.thermoresistible__bu LG--AVSSLNGTLVRPHDIWVGRHP--ELPTSDSTLQATGVVRARIDRVV
MSMEG_4530|M.smegmatis_MC2_155 LG--AVSEINGILVRPHDIRVGRTPEMAIAASDGTAEATGVLRATIDRIV
Mb2419c|M.bovis_AF2122/97 LG--AVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRVV
Rv2397c|M.tuberculosis_H37Rv LG--AVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRVV
MMAR_3715|M.marinum_M LG--AVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRIV
MUL_3658|M.ulcerans_Agy99 LG--AVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRIV
MAV_1785|M.avium_104 LG--AVSTLNGTLVRPHDIRVGRTPEMAVAAEDGTAESTGVARAIVDRVV
MAB_1655|M.abscessus_ATCC_1997 LG--PVAKLNGILVRPHDIRVGRNADLARAAAEGTGETTGVTKATVDRVV
MLBr_01424|M.leprae_Br4923 IGSPGMNLVTLSIVDSSVLLGDWPIRIPREIASAASEVIIGVRPEHFELG
:* : :. :* . : . . : :. .:
Mflv_2681|M.gilvum_PYR-GCK MLGFEVRVELLNAATHTPFTAQITRGDAEALGLREGDTVYVRATRVPSLP
Mvan_3872|M.vanbaalenii_PYR-1 VLGFEVRVELTSAATHTPFTAQITRGDAEALGLKEGDTVYVRATRVPSLP
TH_2424|M.thermoresistible__bu MLGFEVRLELTSATNNAPFTAQITRGDAEALGLRSGDTVYVRATRVPALP
MSMEG_4530|M.smegmatis_MC2_155 MLGFEVRVELTSAASHVPFTAQITRGDAEALGLEEGDTVYVRATRVPHIA
Mb2419c|M.bovis_AF2122/97 VLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPIA
Rv2397c|M.tuberculosis_H37Rv VLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPIA
MMAR_3715|M.marinum_M VLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPIA
MUL_3658|M.ulcerans_Agy99 VLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPIA
MAV_1785|M.avium_104 KLGFEVRVELTSAATGGPFTAQITRGDAEALALREGDTVYVRATRVPPIT
MAB_1655|M.abscessus_ATCC_1997 YLGFEVRVELTSAATGEQFTAQITRGDAEALGLREGDPVYVRVTRVPDVG
MLBr_01424|M.leprae_Br4923 SLGVEVEIDMVEELGADAYLYGRIAGASKVTDQLVVARVDGRNPPAKGSR
**.**.::: . : * ::. * * . .
Mflv_2681|M.gilvum_PYR-GCK T---DDPVAVP-----------------
Mvan_3872|M.vanbaalenii_PYR-1 D---GVPLPVA-----------------
TH_2424|M.thermoresistible__bu TGDVDERVAAR-----------------
MSMEG_4530|M.smegmatis_MC2_155 GEAQVPRTATNNVNADAALAQ-------
Mb2419c|M.bovis_AF2122/97 G---GVSGVDDAGVERVKVTST------
Rv2397c|M.tuberculosis_H37Rv G---GVSGVDDAGVERVKVTST------
MMAR_3715|M.marinum_M G---AAPAVPDDGAGSTPSAAQIGVGGQ
MUL_3658|M.ulcerans_Agy99 G---AAPAVPDDGAGSTPRAAQIGVGGQ
MAV_1785|M.avium_104 AGATTVPAVSRDGADEATLTSA------
MAB_1655|M.abscessus_ATCC_1997 E---LAEVAS------------------
MLBr_01424|M.leprae_Br4923 VRLYTEPGNVHFFGVDGHRIS-------