For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EPLELLAGRVRTADIVDAMGRLHRHRCHLLDLVSPTPGRRLFGPAVTISYFPACSAALPPDCHNFRRLFY QAIERGGEGCVLVLASNGFTEVSLGGGTKLSRAENHRLAGVLADGRLRDFAELEQYDLAVYCRGETTRWG GDTVTPFEANHPVVIGGVAVFPGDYIFADTSGAAVIPADDVRAVLHTATQVVASDAAVIAGIKNEQLDSS *
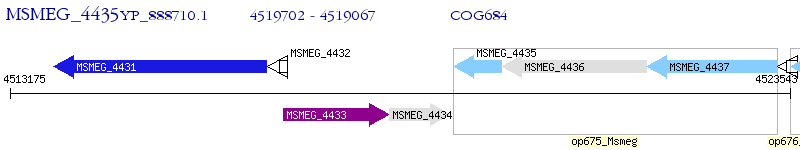
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4435 | - | - | 100% (211) | dimethylmenaquinone methyltransferase |
| M. smegmatis MC2 155 | MSMEG_0707 | - | 4e-11 | 28.10% (121) | hypothetical protein MSMEG_0707 |
| M. smegmatis MC2 155 | MSMEG_0706 | - | 8e-10 | 28.92% (166) | 4-carboxy-4-hydroxy-2-oxoadipate aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3883 | menG | 4e-06 | 27.11% (166) | ribonuclease activity regulator protein RraA |
| M. gilvum PYR-GCK | Mflv_1124 | - | 5e-08 | 27.22% (169) | ribonuclease activity regulator protein RraA |
| M. tuberculosis H37Rv | Rv3853 | menG | 4e-06 | 27.11% (166) | ribonuclease activity regulator protein RraA |
| M. leprae Br4923 | MLBr_00066 | menG | 1e-07 | 26.04% (169) | ribonuclease activity regulator protein RraA |
| M. abscessus ATCC 19977 | MAB_0107c | - | 6e-08 | 29.91% (117) | hypothetical protein MAB_0107c |
| M. marinum M | MMAR_3280 | - | 3e-48 | 48.31% (207) | hypothetical protein MMAR_3280 |
| M. avium 104 | MAV_0174 | - | 2e-08 | 29.41% (170) | ribonuclease activity regulator protein RraA |
| M. thermoresistible (build 8) | TH_1177 | menG | 1e-07 | 28.48% (165) | PROBABLE S-ADENOSYLMETHIONINE:2-DEMETHYLMENAQUINONE |
| M. ulcerans Agy99 | MUL_1349 | - | 8e-49 | 48.79% (207) | hypothetical protein MUL_1349 |
| M. vanbaalenii PYR-1 | Mvan_5682 | - | 2e-07 | 27.38% (168) | ribonuclease activity regulator protein RraA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1124|M.gilvum_PYR-GCK -----MAVTPRPTADLVDEIG--PEVRSCDLQMRQFGGRSEFAGPITTVR
Mvan_5682|M.vanbaalenii_PYR-1 -----MTVTPRPTADLVDEIG--PDVRSCDLQLRQYGGRTDFAGPITTVR
TH_1177|M.thermoresistible__bu -----VIVEPRPTADLVDEIG--PDVRSCDLQLQQYGGRAQFAGPISTVR
Mb3883|M.bovis_AF2122/97 -----MAISFRPTADLVDDIG--PDVRSCDLQFRQFGGRSQFAGPISTVR
Rv3853|M.tuberculosis_H37Rv -----MAISFRPTADLVDDIG--PDVRSCDLQFRQFGGRSQFAGPISTVR
MLBr_00066|M.leprae_Br4923 -----MIVSFRPTADLVDSIG--VDVRSCDLQFRQFGGCSEFAGPISTVR
MAV_0174|M.avium_104 -----MTVSFRPTADLVDDIG--PDVRSCDLQFRQLGGRTEFAGPISTVR
MAB_0107c|M.abscessus_ATCC_199 --MTTAPITPRPTADLGDELG--IDMASCDLQLNQYGARPVFAGVITTVR
MMAR_3280|M.marinum_M MQLTELVDG-LGVADVVDAMMMTYAHRAHIIELDSPDPDRVLVGPAVTIS
MUL_1349|M.ulcerans_Agy99 MQLTELVDG-LGVADVVDAMMMTYAHRAHIIELDSPDPDRVLVGPAVTIS
MSMEG_4435|M.smegmatis_MC2_155 MEPLELLAGRVRTADIVDAMGRLHRHRCHLLDLVSPTPGRRLFGPAVTIS
.**: * : . ::: . : * *:
Mflv_1124|M.gilvum_PYR-GCK CFEDN-------------ALLKSVLSEPGDGGVLVVDGDGSVHTALVGDV
Mvan_5682|M.vanbaalenii_PYR-1 CFQDN-------------ALLKSVLSEPGDGGVLVIDGDGSVHTALVGDV
TH_1177|M.thermoresistible__bu CFQDN-------------ALLKSVLSEPGNGGVLVIDGDGSLHTALVGDV
Mb3883|M.bovis_AF2122/97 CFQDN-------------ALLKSVLSQPSAGGVLVIDGAGSLHTALVGDV
Rv3853|M.tuberculosis_H37Rv CFQDN-------------ALLKSVLSQPSAGGVLVIDGAGSLHTALVGDV
MLBr_00066|M.leprae_Br4923 CFQDN-------------ALLKSVLSQTSAGGVLVVDGAGSLHTALVGDV
MAV_0174|M.avium_104 CFQDN-------------ALLKSVLSEPGGGGVLVVDGGGSLHTALVGDV
MAB_0107c|M.abscessus_ATCC_199 CHHDN-------------ALLKSVLSEPSSGGVLVIDGGGSLHCALVGDV
MMAR_3280|M.marinum_M FLPVRKDLMDPEKHSLGPAIYRAVAKHDPAGAVLVMASNGYHQTSLGGGT
MUL_1349|M.ulcerans_Agy99 FLPVRKDLMDPEKHSLGPAIYRAVARHDPAGAVLVMASNGYHQTSLGGGT
MSMEG_4435|M.smegmatis_MC2_155 YFPACSAALPPDCHNFRRLFYQAIERGG-EGCVLVLASNGFTEVSLGGGT
: ::: * ***: . * . :* *..
Mflv_1124|M.gilvum_PYR-GCK IAELGRSNGWSGLIINGAVRDASTLRTLDIGIKALGTNPR-KSSKTGDGV
Mvan_5682|M.vanbaalenii_PYR-1 IAELGRSNGWSGLIINGAVRDASTLRTLDIGIKALGTNPR-KSTKTGEGI
TH_1177|M.thermoresistible__bu IAELARGNGWAGLIINGAVRDSATLRTLDIGIKALGTNPR-KSTKTGAGQ
Mb3883|M.bovis_AF2122/97 IAELARSTGWTGLIVHGAVRDAAALRGIDIGIKALGTNPR-KSTKTGAGE
Rv3853|M.tuberculosis_H37Rv IAELARSTGWTGLIVHGAVRDAAALRGIDIGIKALGTNPR-KSTKTGAGE
MLBr_00066|M.leprae_Br4923 IAELAHSNGWAGLIVNGAVRDAAALRGIDIGIKALGTNPR-KSTKIGTGE
MAV_0174|M.avium_104 IAELARANGWAGLIVNGAVRDSAALRGMDIGVKALGTNPR-KSTKTGAGE
MAB_0107c|M.abscessus_ATCC_199 IAGLAVASGWSGLVINGVVRDSAELATMDIGIKALGTNPR-KGHKTGAGE
MMAR_3280|M.marinum_M KLSRVENLGMAGILADGMLRDFEELSTYNFATYCHGETARGGGNEIQPYL
MUL_1349|M.ulcerans_Agy99 KLSRVENLGMAGILADGMLRDFEELSTYNFATYCHGETARGGGNEIQPYL
MSMEG_4435|M.smegmatis_MC2_155 KLSRAENHRLAGVLADGRLRDFAELEQYDLAVYCRGETTRWGGDTVTPFE
:*:: .* :** * ::. . * ..* .
Mflv_1124|M.gilvum_PYR-GCK RDAVVEFGGVVFTPGDIAYSDDDGIVVVTAD-------------------
Mvan_5682|M.vanbaalenii_PYR-1 RDEAVEFGGVVFTPGDIAYSDDDGIVVIAAG-------------------
TH_1177|M.thermoresistible__bu RDVAVEFGGVTFVPGEIAYADEDGIVVXXGGNGGRGGSVILVVDPQVHTL
Mb3883|M.bovis_AF2122/97 RDVEITLGGVTFVPGDIAYSDDDGIIVV----------------------
Rv3853|M.tuberculosis_H37Rv RDVEITLGGVTFVPGDIAYSDDDGIIVV----------------------
MLBr_00066|M.leprae_Br4923 RHVEVNLGGVTFVPGEVVYSDDDGIVVV----------------------
MAV_0174|M.avium_104 RDVDITLGGVTFTPGDIAYSDDDGIVVVAAGD------------------
MAB_0107c|M.abscessus_ATCC_199 RDVVVSFGGIDFTPGAICYADHDGVVVVPA--------------------
MMAR_3280|M.marinum_M ADVPVAIGGVTVVPGDVIFAKGSTAVVIPGDQAEAILTKAHNIMKKMDQV
MUL_1349|M.ulcerans_Agy99 ADVPVAIGGVTVVPGDVIFAKGSTAVVIPGDQAEAILTKAHNIMKKMDQV
MSMEG_4435|M.smegmatis_MC2_155 ANHPVVIGGVAVFPGDYIFADTSGAAVIPADDVRAVLHTATQVVASDAAV
. : :**: . ** ::. . *
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu LDFHFHPHVVAPSGKPGQGSNRDGAAGADLEVKVPDGTVVLDEHGNLLAD
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M KESLKSEDPETVLRQGSGEL------------------------------
MUL_1349|M.ulcerans_Agy99 KESLKSEDPETVLRQGSGEL------------------------------
MSMEG_4435|M.smegmatis_MC2_155 IAGIKNEQLDSS--------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu LTGAGTRFVAAEGGRGGLGNAALASRARKAPGFALLGEPGQARDLVLELK
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu TVADVGLIGYPSAGKSSLVSTISAAKPKIADYPFTTLTPNLGVVSAGDET
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu FTVADVPGLIPGASEGRGLGLDFLRHIERCAVLVHVVDCATLELGRDPVS
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu DIETLENELAAYTPTLQGDSELDDLTERPRAVVLNKIDVPEARELAEFVR
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu DEIVQRFGWPVFLVSTATREGLRPLVFALWEMMKQYRASQPEVAPRRPVI
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu RPVPVDETGFTVHPDGEGGFVVRGVRPERWVTQTDFNNDEAVGYLADRLA
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu RLGVEEELLQLGAEPGCEVTIGDMTFEWEPQTPAGVDDVPLSGRGTDARL
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4435|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK -------------------------
Mvan_5682|M.vanbaalenii_PYR-1 -------------------------
TH_1177|M.thermoresistible__bu EQSERVTAAERKAARRARREPGGEP
Mb3883|M.bovis_AF2122/97 -------------------------
Rv3853|M.tuberculosis_H37Rv -------------------------
MLBr_00066|M.leprae_Br4923 -------------------------
MAV_0174|M.avium_104 -------------------------
MAB_0107c|M.abscessus_ATCC_199 -------------------------
MMAR_3280|M.marinum_M -------------------------
MUL_1349|M.ulcerans_Agy99 -------------------------
MSMEG_4435|M.smegmatis_MC2_155 -------------------------