For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSTRTPTERLSALPTAAVSDALDARGLPGSLAGIGRVTGSTLTGTAFTVRYEPVAGDDGTVGDFLDDIPR GSAVVIDNAGRLDCTVWGGIMSRVAAARGIAGTVINGVCRDVTASQEVGYSLWAAGRFMRTGKDRVRCVA VQTPVVIDDVTINPGDYVHADADGVVVVPAPHIDATLDLAEHIERIETVIVETTIAGTPLAEARRRFGYH TLQTRSAG*
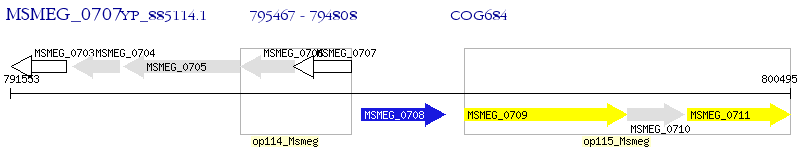
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0707 | - | - | 100% (219) | hypothetical protein MSMEG_0707 |
| M. smegmatis MC2 155 | MSMEG_0706 | - | 9e-15 | 34.42% (154) | 4-carboxy-4-hydroxy-2-oxoadipate aldolase |
| M. smegmatis MC2 155 | MSMEG_4435 | - | 5e-11 | 28.10% (121) | dimethylmenaquinone methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3883 | menG | 2e-08 | 26.79% (112) | ribonuclease activity regulator protein RraA |
| M. gilvum PYR-GCK | Mflv_1124 | - | 7e-08 | 27.97% (118) | ribonuclease activity regulator protein RraA |
| M. tuberculosis H37Rv | Rv3853 | menG | 2e-08 | 26.79% (112) | ribonuclease activity regulator protein RraA |
| M. leprae Br4923 | MLBr_00066 | menG | 2e-10 | 28.57% (161) | ribonuclease activity regulator protein RraA |
| M. abscessus ATCC 19977 | MAB_0107c | - | 1e-10 | 34.48% (116) | hypothetical protein MAB_0107c |
| M. marinum M | MMAR_3280 | - | 6e-09 | 25.00% (140) | hypothetical protein MMAR_3280 |
| M. avium 104 | MAV_0174 | - | 2e-09 | 28.95% (114) | ribonuclease activity regulator protein RraA |
| M. thermoresistible (build 8) | TH_1177 | menG | 2e-08 | 31.30% (115) | PROBABLE S-ADENOSYLMETHIONINE:2-DEMETHYLMENAQUINONE |
| M. ulcerans Agy99 | MUL_1349 | - | 4e-09 | 25.00% (140) | hypothetical protein MUL_1349 |
| M. vanbaalenii PYR-1 | Mvan_5682 | - | 1e-07 | 27.61% (163) | ribonuclease activity regulator protein RraA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1124|M.gilvum_PYR-GCK --------MAVTPRPTADLVDEIGP--EVRSCDLQMRQFGGRSEFAGPIT
Mvan_5682|M.vanbaalenii_PYR-1 --------MTVTPRPTADLVDEIGP--DVRSCDLQLRQYGGRTDFAGPIT
TH_1177|M.thermoresistible__bu --------VIVEPRPTADLVDEIGP--DVRSCDLQLQQYGGRAQFAGPIS
Mb3883|M.bovis_AF2122/97 --------MAISFRPTADLVDDIGP--DVRSCDLQFRQFGGRSQFAGPIS
Rv3853|M.tuberculosis_H37Rv --------MAISFRPTADLVDDIGP--DVRSCDLQFRQFGGRSQFAGPIS
MLBr_00066|M.leprae_Br4923 --------MIVSFRPTADLVDSIGV--DVRSCDLQFRQFGGCSEFAGPIS
MAV_0174|M.avium_104 --------MTVSFRPTADLVDDIGP--DVRSCDLQFRQLGGRTEFAGPIS
MAB_0107c|M.abscessus_ATCC_199 -----MTTAPITPRPTADLGDELGI--DMASCDLQLNQYGARPVFAGVIT
MMAR_3280|M.marinum_M ----MQLTELVDGLGVADVVDAMMMTYAHRAHIIELDSPDPDRVLVGPAV
MUL_1349|M.ulcerans_Agy99 ----MQLTELVDGLGVADVVDAMMMTYAHRAHIIELDSPDPDRVLVGPAV
MSMEG_0707|M.smegmatis_MC2_155 MTSTRTPTERLSALPTAAVSDALDAR-GLPGSLAGIGRVTG-STLTGTAF
: .* : * : . : :.*
Mflv_1124|M.gilvum_PYR-GCK TVRCFEDN-------------ALLKSVLSEPGDGGVLVVDGDGSVHTALV
Mvan_5682|M.vanbaalenii_PYR-1 TVRCFQDN-------------ALLKSVLSEPGDGGVLVIDGDGSVHTALV
TH_1177|M.thermoresistible__bu TVRCFQDN-------------ALLKSVLSEPGNGGVLVIDGDGSLHTALV
Mb3883|M.bovis_AF2122/97 TVRCFQDN-------------ALLKSVLSQPSAGGVLVIDGAGSLHTALV
Rv3853|M.tuberculosis_H37Rv TVRCFQDN-------------ALLKSVLSQPSAGGVLVIDGAGSLHTALV
MLBr_00066|M.leprae_Br4923 TVRCFQDN-------------ALLKSVLSQTSAGGVLVVDGAGSLHTALV
MAV_0174|M.avium_104 TVRCFQDN-------------ALLKSVLSEPGGGGVLVVDGGGSLHTALV
MAB_0107c|M.abscessus_ATCC_199 TVRCHHDN-------------ALLKSVLSEPSSGGVLVIDGGGSLHCALV
MMAR_3280|M.marinum_M TISFLPVRKDLMDPEKHSLGPAIYRAVAKHDPAGAVLVMASNGYHQTSLG
MUL_1349|M.ulcerans_Agy99 TISFLPVRKDLMDPEKHSLGPAIYRAVARHDPAGAVLVMASNGYHQTSLG
MSMEG_0707|M.smegmatis_MC2_155 TVRYEPVAGD----------DGTVGDFLDDIPRGSAVVIDNAGRLDCTVW
*: . . . *..:*: . * . ::
Mflv_1124|M.gilvum_PYR-GCK GDVIAELGRSNGWSGLIINGAVRDASTLRTLDIGIKALGTNPRKS-SKTG
Mvan_5682|M.vanbaalenii_PYR-1 GDVIAELGRSNGWSGLIINGAVRDASTLRTLDIGIKALGTNPRKS-TKTG
TH_1177|M.thermoresistible__bu GDVIAELARGNGWAGLIINGAVRDSATLRTLDIGIKALGTNPRKS-TKTG
Mb3883|M.bovis_AF2122/97 GDVIAELARSTGWTGLIVHGAVRDAAALRGIDIGIKALGTNPRKS-TKTG
Rv3853|M.tuberculosis_H37Rv GDVIAELARSTGWTGLIVHGAVRDAAALRGIDIGIKALGTNPRKS-TKTG
MLBr_00066|M.leprae_Br4923 GDVIAELAHSNGWAGLIVNGAVRDAAALRGIDIGIKALGTNPRKS-TKIG
MAV_0174|M.avium_104 GDVIAELARANGWAGLIVNGAVRDSAALRGMDIGVKALGTNPRKS-TKTG
MAB_0107c|M.abscessus_ATCC_199 GDVIAGLAVASGWSGLVINGVVRDSAELATMDIGIKALGTNPRKG-HKTG
MMAR_3280|M.marinum_M GGTKLSRVENLGMAGILADGMLRDFEELSTYNFATYCHGETARGGGNEIQ
MUL_1349|M.ulcerans_Agy99 GGTKLSRVENLGMAGILADGMLRDFEELSTYNFATYCHGETARGGGNEIQ
MSMEG_0707|M.smegmatis_MC2_155 GGIMSRVAAARGIAGTVINGVCRDVTASQEVGYSLWAAGRFMRTGKDRVR
*. * :* : .* ** . . . * * . .
Mflv_1124|M.gilvum_PYR-GCK DGVRDAVVEFGGVVFTPGDIAYSDDDGIVVVTAD----------------
Mvan_5682|M.vanbaalenii_PYR-1 EGIRDEAVEFGGVVFTPGDIAYSDDDGIVVIAAG----------------
TH_1177|M.thermoresistible__bu AGQRDVAVEFGGVTFVPGEIAYADEDGIVVXXGGNGGRGGSVILVVDPQV
Mb3883|M.bovis_AF2122/97 AGERDVEITLGGVTFVPGDIAYSDDDGIIVV-------------------
Rv3853|M.tuberculosis_H37Rv AGERDVEITLGGVTFVPGDIAYSDDDGIIVV-------------------
MLBr_00066|M.leprae_Br4923 TGERHVEVNLGGVTFVPGEVVYSDDDGIVVV-------------------
MAV_0174|M.avium_104 AGERDVDITLGGVTFTPGDIAYSDDDGIVVVAAGD---------------
MAB_0107c|M.abscessus_ATCC_199 AGERDVVVSFGGIDFTPGAICYADHDGVVVVPA-----------------
MMAR_3280|M.marinum_M PYLADVPVAIGGVTVVPGDVIFAKGSTAVVIPGDQAEAILTKAHNIMKKM
MUL_1349|M.ulcerans_Agy99 PYLADVPVAIGGVTVVPGDVIFAKGSTAVVIPGDQAEAILTKAHNIMKKM
MSMEG_0707|M.smegmatis_MC2_155 CVAVQTPVVIDDVTINPGDYVHADADGVVVVPAPHIDATLDLAEHIERIE
. : :..: . ** .:. . :*
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu HTLLDFHFHPHVVAPSGKPGQGSNRDGAAGADLEVKVPDGTVVLDEHGNL
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M DQVKESLKSEDPETVLRQGSGEL---------------------------
MUL_1349|M.ulcerans_Agy99 DQVKESLKSEDPETVLRQGSGEL---------------------------
MSMEG_0707|M.smegmatis_MC2_155 TVIVETTIAGTPLAEARRRFGYHTLQTRSAG-------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu LADLTGAGTRFVAAEGGRGGLGNAALASRARKAPGFALLGEPGQARDLVL
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu ELKTVADVGLIGYPSAGKSSLVSTISAAKPKIADYPFTTLTPNLGVVSAG
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu DETFTVADVPGLIPGASEGRGLGLDFLRHIERCAVLVHVVDCATLELGRD
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu PVSDIETLENELAAYTPTLQGDSELDDLTERPRAVVLNKIDVPEARELAE
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu FVRDEIVQRFGWPVFLVSTATREGLRPLVFALWEMMKQYRASQPEVAPRR
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu PVIRPVPVDETGFTVHPDGEGGFVVRGVRPERWVTQTDFNNDEAVGYLAD
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu RLARLGVEEELLQLGAEPGCEVTIGDMTFEWEPQTPAGVDDVPLSGRGTD
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3280|M.marinum_M --------------------------------------------------
MUL_1349|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0707|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK ----------------------------
Mvan_5682|M.vanbaalenii_PYR-1 ----------------------------
TH_1177|M.thermoresistible__bu ARLEQSERVTAAERKAARRARREPGGEP
Mb3883|M.bovis_AF2122/97 ----------------------------
Rv3853|M.tuberculosis_H37Rv ----------------------------
MLBr_00066|M.leprae_Br4923 ----------------------------
MAV_0174|M.avium_104 ----------------------------
MAB_0107c|M.abscessus_ATCC_199 ----------------------------
MMAR_3280|M.marinum_M ----------------------------
MUL_1349|M.ulcerans_Agy99 ----------------------------
MSMEG_0707|M.smegmatis_MC2_155 ----------------------------