For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVTPRPTADLVDEIGPEVRSCDLQMRQFGGRSEFAGPITTVRCFEDNALLKSVLSEPGDGGVLVVDGDGS VHTALVGDVIAELGRSNGWSGLIINGAVRDASTLRTLDIGIKALGTNPRKSSKTGDGVRDAVVEFGGVVF TPGDIAYSDDDGIVVVTAD*
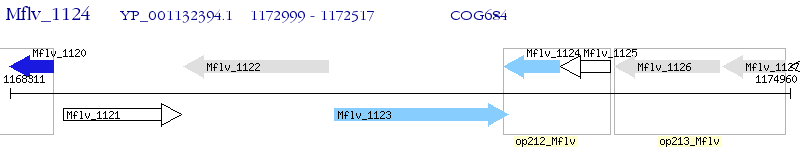
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1124 | - | - | 100% (160) | ribonuclease activity regulator protein RraA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3883 | menG | 7e-69 | 77.07% (157) | ribonuclease activity regulator protein RraA |
| M. tuberculosis H37Rv | Rv3853 | menG | 7e-69 | 77.07% (157) | ribonuclease activity regulator protein RraA |
| M. leprae Br4923 | MLBr_00066 | menG | 1e-64 | 76.13% (155) | ribonuclease activity regulator protein RraA |
| M. abscessus ATCC 19977 | MAB_0107c | - | 5e-60 | 69.43% (157) | hypothetical protein MAB_0107c |
| M. marinum M | MMAR_5403 | menG | 3e-70 | 79.22% (154) | s-adenosylmethionine:2-demethylmenaquinone |
| M. avium 104 | MAV_0174 | - | 1e-70 | 78.62% (159) | ribonuclease activity regulator protein RraA |
| M. smegmatis MC2 155 | MSMEG_6439 | - | 1e-66 | 73.42% (158) | ribonuclease activity regulator protein RraA |
| M. thermoresistible (build 8) | TH_1177 | menG | 2e-73 | 82.47% (154) | PROBABLE S-ADENOSYLMETHIONINE:2-DEMETHYLMENAQUINONE |
| M. ulcerans Agy99 | MUL_5025 | menG | 7e-70 | 78.57% (154) | ribonuclease activity regulator protein RraA |
| M. vanbaalenii PYR-1 | Mvan_5682 | - | 3e-82 | 89.94% (159) | ribonuclease activity regulator protein RraA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3883|M.bovis_AF2122/97 --MA-ISFRPTADLVDDIGPDVRSCDLQFRQFGGRSQFAGPISTVRCFQD
Rv3853|M.tuberculosis_H37Rv --MA-ISFRPTADLVDDIGPDVRSCDLQFRQFGGRSQFAGPISTVRCFQD
MMAR_5403|M.marinum_M --MGNVAFRPTADLVDDIGPDVRSCDLQFRQFGGRAEFAGPISTVRCFQD
MUL_5025|M.ulcerans_Agy99 --MGNVAFRPTADLVDDIGPDVRSCDLQFRQFGGHAEFAGPISTVRCFQD
MLBr_00066|M.leprae_Br4923 ---MIVSFRPTADLVDSIGVDVRSCDLQFRQFGGCSEFAGPISTVRCFQD
MAV_0174|M.avium_104 ---MTVSFRPTADLVDDIGPDVRSCDLQFRQLGGRTEFAGPISTVRCFQD
Mflv_1124|M.gilvum_PYR-GCK ---MAVTPRPTADLVDEIGPEVRSCDLQMRQFGGRSEFAGPITTVRCFED
Mvan_5682|M.vanbaalenii_PYR-1 ---MTVTPRPTADLVDEIGPDVRSCDLQLRQYGGRTDFAGPITTVRCFQD
TH_1177|M.thermoresistible__bu ---VIVEPRPTADLVDEIGPDVRSCDLQLQQYGGRAQFAGPISTVRCFQD
MSMEG_6439|M.smegmatis_MC2_155 ---MSIEPRATADLVDEIYPDVRSCDLQLQNYGGKIMFAGPVTTVRCFQD
MAB_0107c|M.abscessus_ATCC_199 MTTAPITPRPTADLGDELGIDMASCDLQLNQYGARPVFAGVITTVRCHHD
: *.**** *.: :: *****:.: *. *** ::****..*
Mb3883|M.bovis_AF2122/97 NALLKSVLSQPSAGGVLVIDGAGSLHTALVGDVIAELARSTGWTGLIVHG
Rv3853|M.tuberculosis_H37Rv NALLKSVLSQPSAGGVLVIDGAGSLHTALVGDVIAELARSTGWTGLIVHG
MMAR_5403|M.marinum_M NALLKSVLSQPGEGGVLVIDGAGSLHTALVGDVIAELARSNGWAGLVVHG
MUL_5025|M.ulcerans_Agy99 NALLKSVLSQPGEGGVLVIDGAGSLHTALVGDVIAELARSNGWTGLVVHG
MLBr_00066|M.leprae_Br4923 NALLKSVLSQTSAGGVLVVDGAGSLHTALVGDVIAELAHSNGWAGLIVNG
MAV_0174|M.avium_104 NALLKSVLSEPGGGGVLVVDGGGSLHTALVGDVIAELARANGWAGLIVNG
Mflv_1124|M.gilvum_PYR-GCK NALLKSVLSEPGDGGVLVVDGDGSVHTALVGDVIAELGRSNGWSGLIING
Mvan_5682|M.vanbaalenii_PYR-1 NALLKSVLSEPGDGGVLVIDGDGSVHTALVGDVIAELGRSNGWSGLIING
TH_1177|M.thermoresistible__bu NALLKSVLSEPGNGGVLVIDGDGSLHTALVGDVIAELARGNGWAGLIING
MSMEG_6439|M.smegmatis_MC2_155 NALLKSILSTPGEGAVLVVDGAGSLHTALVGDVIAGLAADNGWSGVIVNG
MAB_0107c|M.abscessus_ATCC_199 NALLKSVLSEPSSGGVLVIDGGGSLHCALVGDVIAGLAVASGWSGLVING
******:** .. *.***:** **:* ******** *. .**:*::::*
Mb3883|M.bovis_AF2122/97 AVRDAAALRGIDIGIKALGTNPRKSTKTGAGERDVEITLGGVTFVPGDIA
Rv3853|M.tuberculosis_H37Rv AVRDAAALRGIDIGIKALGTNPRKSTKTGAGERDVEITLGGVTFVPGDIA
MMAR_5403|M.marinum_M AVRDSAALRGIDIGIKALGTNPRKSTKTGSGERDAEITLGGVTFAPGEIA
MUL_5025|M.ulcerans_Agy99 AVRDSAALRGIDIGIKALGTNPRKSTKTGSGERDAEITLGGVTFVPGEIA
MLBr_00066|M.leprae_Br4923 AVRDAAALRGIDIGIKALGTNPRKSTKIGTGERHVEVNLGGVTFVPGEVV
MAV_0174|M.avium_104 AVRDSAALRGMDIGVKALGTNPRKSTKTGAGERDVDITLGGVTFTPGDIA
Mflv_1124|M.gilvum_PYR-GCK AVRDASTLRTLDIGIKALGTNPRKSSKTGDGVRDAVVEFGGVVFTPGDIA
Mvan_5682|M.vanbaalenii_PYR-1 AVRDASTLRTLDIGIKALGTNPRKSTKTGEGIRDEAVEFGGVVFTPGDIA
TH_1177|M.thermoresistible__bu AVRDSATLRTLDIGIKALGTNPRKSTKTGAGQRDVAVEFGGVTFVPGEIA
MSMEG_6439|M.smegmatis_MC2_155 AVRDAAALRTIDVGIKALGTNPRKGTKTGEGERDVEVSFGGVTFTPGDIA
MAB_0107c|M.abscessus_ATCC_199 VVRDSAELATMDIGIKALGTNPRKGHKTGAGERDVVVSFGGIDFTPGAIC
.***:: * :*:*:*********. * * * *. : :**: *.** :
Mb3883|M.bovis_AF2122/97 YSDDDGIIVV----------------------------------------
Rv3853|M.tuberculosis_H37Rv YSDDDGIIVV----------------------------------------
MMAR_5403|M.marinum_M YSDDDGIVVVAAVATES---------------------------------
MUL_5025|M.ulcerans_Agy99 YSDDDGIVVVAAVATES---------------------------------
MLBr_00066|M.leprae_Br4923 YSDDDGIVVV----------------------------------------
MAV_0174|M.avium_104 YSDDDGIVVVAAGD------------------------------------
Mflv_1124|M.gilvum_PYR-GCK YSDDDGIVVVTAD-------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 YSDDDGIVVIAAG-------------------------------------
TH_1177|M.thermoresistible__bu YADEDGIVVXXGGNGGRGGSVILVVDPQVHTLLDFHFHPHVVAPSGKPGQ
MSMEG_6439|M.smegmatis_MC2_155 YCDEDGIVVVTP--------------------------------------
MAB_0107c|M.abscessus_ATCC_199 YADHDGVVVVPA--------------------------------------
*.*.**::*
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu GSNRDGAAGADLEVKVPDGTVVLDEHGNLLADLTGAGTRFVAAEGGRGGL
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu GNAALASRARKAPGFALLGEPGQARDLVLELKTVADVGLIGYPSAGKSSL
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu VSTISAAKPKIADYPFTTLTPNLGVVSAGDETFTVADVPGLIPGASEGRG
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu LGLDFLRHIERCAVLVHVVDCATLELGRDPVSDIETLENELAAYTPTLQG
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu DSELDDLTERPRAVVLNKIDVPEARELAEFVRDEIVQRFGWPVFLVSTAT
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu REGLRPLVFALWEMMKQYRASQPEVAPRRPVIRPVPVDETGFTVHPDGEG
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu GFVVRGVRPERWVTQTDFNNDEAVGYLADRLARLGVEEELLQLGAEPGCE
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1177|M.thermoresistible__bu VTIGDMTFEWEPQTPAGVDDVPLSGRGTDARLEQSERVTAAERKAARRAR
MSMEG_6439|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 -------
Rv3853|M.tuberculosis_H37Rv -------
MMAR_5403|M.marinum_M -------
MUL_5025|M.ulcerans_Agy99 -------
MLBr_00066|M.leprae_Br4923 -------
MAV_0174|M.avium_104 -------
Mflv_1124|M.gilvum_PYR-GCK -------
Mvan_5682|M.vanbaalenii_PYR-1 -------
TH_1177|M.thermoresistible__bu REPGGEP
MSMEG_6439|M.smegmatis_MC2_155 -------
MAB_0107c|M.abscessus_ATCC_199 -------