For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDLFTNHTSATLLEAAQRDVALDPAIAPVWRGARVAGPAFTVQGVGGDNLALHNAVCKCPPGHVLVVDAG GQAFGHWGEILAVAAMSRGIRGLVIDGAVRDSVEQENLGFPVFSRGLAIRGTGKSYPGVLGRPVRVGGVK VRTGDLVVGDADGVVSLPAEQVPEILERADARIRKEEQVISALRQGATTIDLYQFDPTGLGN*
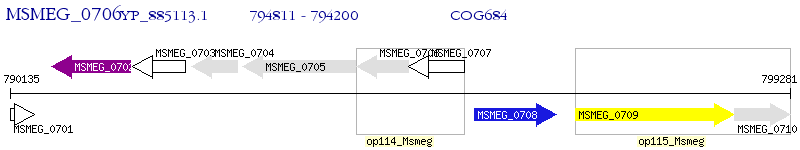
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0706 | - | - | 100% (203) | 4-carboxy-4-hydroxy-2-oxoadipate aldolase |
| M. smegmatis MC2 155 | MSMEG_0707 | - | 8e-15 | 34.42% (154) | hypothetical protein MSMEG_0707 |
| M. smegmatis MC2 155 | MSMEG_4435 | - | 8e-10 | 28.92% (166) | dimethylmenaquinone methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3883 | menG | 1e-09 | 30.65% (124) | ribonuclease activity regulator protein RraA |
| M. gilvum PYR-GCK | Mflv_1124 | - | 3e-11 | 34.92% (126) | ribonuclease activity regulator protein RraA |
| M. tuberculosis H37Rv | Rv3853 | menG | 1e-09 | 30.65% (124) | ribonuclease activity regulator protein RraA |
| M. leprae Br4923 | MLBr_00066 | menG | 3e-10 | 33.88% (121) | ribonuclease activity regulator protein RraA |
| M. abscessus ATCC 19977 | MAB_0107c | - | 4e-10 | 34.51% (113) | hypothetical protein MAB_0107c |
| M. marinum M | MMAR_5403 | menG | 2e-10 | 32.81% (128) | s-adenosylmethionine:2-demethylmenaquinone |
| M. avium 104 | MAV_0174 | - | 4e-11 | 34.40% (125) | ribonuclease activity regulator protein RraA |
| M. thermoresistible (build 8) | TH_1177 | menG | 5e-10 | 33.87% (124) | PROBABLE S-ADENOSYLMETHIONINE:2-DEMETHYLMENAQUINONE |
| M. ulcerans Agy99 | MUL_5025 | menG | 2e-10 | 32.81% (128) | ribonuclease activity regulator protein RraA |
| M. vanbaalenii PYR-1 | Mvan_5682 | - | 9e-11 | 33.60% (125) | ribonuclease activity regulator protein RraA |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5403|M.marinum_M --MGNVAFRPTADLVDDIGPDVRSCDLQFRQFGGRAEFAGPISTVR-CFQ
MUL_5025|M.ulcerans_Agy99 --MGNVAFRPTADLVDDIGPDVRSCDLQFRQFGGHAEFAGPISTVR-CFQ
Mb3883|M.bovis_AF2122/97 --MA-ISFRPTADLVDDIGPDVRSCDLQFRQFGGRSQFAGPISTVR-CFQ
Rv3853|M.tuberculosis_H37Rv --MA-ISFRPTADLVDDIGPDVRSCDLQFRQFGGRSQFAGPISTVR-CFQ
MLBr_00066|M.leprae_Br4923 ---MIVSFRPTADLVDSIGVDVRSCDLQFRQFGGCSEFAGPISTVR-CFQ
MAV_0174|M.avium_104 ---MTVSFRPTADLVDDIGPDVRSCDLQFRQLGGRTEFAGPISTVR-CFQ
TH_1177|M.thermoresistible__bu ---VIVEPRPTADLVDEIGPDVRSCDLQLQQYGGRAQFAGPISTVR-CFQ
Mflv_1124|M.gilvum_PYR-GCK ---MAVTPRPTADLVDEIGPEVRSCDLQMRQFGGRSEFAGPITTVR-CFE
Mvan_5682|M.vanbaalenii_PYR-1 ---MTVTPRPTADLVDEIGPDVRSCDLQLRQYGGRTDFAGPITTVR-CFQ
MAB_0107c|M.abscessus_ATCC_199 MTTAPITPRPTADLGDELGIDMASCDLQLNQYGARPVFAGVITTVR-CHH
MSMEG_0706|M.smegmatis_MC2_155 -MSDLFTNHTSATLLEAAQRDVALDPAIAPVWRG-ARVAGPAFTVQGVGG
. :.:* * : :: . . .** **:
MMAR_5403|M.marinum_M DNALLKSVLSQPGEGGVLVIDGAGSLHTALVGDVIAELARSNGWAGLVVH
MUL_5025|M.ulcerans_Agy99 DNALLKSVLSQPGEGGVLVIDGAGSLHTALVGDVIAELARSNGWTGLVVH
Mb3883|M.bovis_AF2122/97 DNALLKSVLSQPSAGGVLVIDGAGSLHTALVGDVIAELARSTGWTGLIVH
Rv3853|M.tuberculosis_H37Rv DNALLKSVLSQPSAGGVLVIDGAGSLHTALVGDVIAELARSTGWTGLIVH
MLBr_00066|M.leprae_Br4923 DNALLKSVLSQTSAGGVLVVDGAGSLHTALVGDVIAELAHSNGWAGLIVN
MAV_0174|M.avium_104 DNALLKSVLSEPGGGGVLVVDGGGSLHTALVGDVIAELARANGWAGLIVN
TH_1177|M.thermoresistible__bu DNALLKSVLSEPGNGGVLVIDGDGSLHTALVGDVIAELARGNGWAGLIIN
Mflv_1124|M.gilvum_PYR-GCK DNALLKSVLSEPGDGGVLVVDGDGSVHTALVGDVIAELGRSNGWSGLIIN
Mvan_5682|M.vanbaalenii_PYR-1 DNALLKSVLSEPGDGGVLVIDGDGSVHTALVGDVIAELGRSNGWSGLIIN
MAB_0107c|M.abscessus_ATCC_199 DNALLKSVLSEPSSGGVLVIDGGGSLHCALVGDVIAGLAVASGWSGLVIN
MSMEG_0706|M.smegmatis_MC2_155 DNLALHNAVCKCPPGHVLVVDAGG-QAFGHWGEILAVAAMSRGIRGLVID
** *:..:.: * ***:*. * . *:::* . . * **::.
MMAR_5403|M.marinum_M GAVRDSAALRGIDIGIKALGTNPRKSTKTGSGERDAEITLGGVTFAPGEI
MUL_5025|M.ulcerans_Agy99 GAVRDSAALRGIDIGIKALGTNPRKSTKTGSGERDAEITLGGVTFVPGEI
Mb3883|M.bovis_AF2122/97 GAVRDAAALRGIDIGIKALGTNPRKSTKTGAGERDVEITLGGVTFVPGDI
Rv3853|M.tuberculosis_H37Rv GAVRDAAALRGIDIGIKALGTNPRKSTKTGAGERDVEITLGGVTFVPGDI
MLBr_00066|M.leprae_Br4923 GAVRDAAALRGIDIGIKALGTNPRKSTKIGTGERHVEVNLGGVTFVPGEV
MAV_0174|M.avium_104 GAVRDSAALRGMDIGVKALGTNPRKSTKTGAGERDVDITLGGVTFTPGDI
TH_1177|M.thermoresistible__bu GAVRDSATLRTLDIGIKALGTNPRKSTKTGAGQRDVAVEFGGVTFVPGEI
Mflv_1124|M.gilvum_PYR-GCK GAVRDASTLRTLDIGIKALGTNPRKSSKTGDGVRDAVVEFGGVVFTPGDI
Mvan_5682|M.vanbaalenii_PYR-1 GAVRDASTLRTLDIGIKALGTNPRKSTKTGEGIRDEAVEFGGVVFTPGDI
MAB_0107c|M.abscessus_ATCC_199 GVVRDSAELATMDIGIKALGTNPRKGHKTGAGERDVVVSFGGIDFTPGAI
MSMEG_0706|M.smegmatis_MC2_155 GAVRDSVEQENLGFPVFSRGLAIRGTGKSYPGVLGRPVRVGGVKVRTGDL
*.***: :.: : : * * * * : .**: . .* :
MMAR_5403|M.marinum_M AYSDDDGIVVVAAVATES--------------------------------
MUL_5025|M.ulcerans_Agy99 AYSDDDGIVVVAAVATES--------------------------------
Mb3883|M.bovis_AF2122/97 AYSDDDGIIVV---------------------------------------
Rv3853|M.tuberculosis_H37Rv AYSDDDGIIVV---------------------------------------
MLBr_00066|M.leprae_Br4923 VYSDDDGIVVV---------------------------------------
MAV_0174|M.avium_104 AYSDDDGIVVVAAGD-----------------------------------
TH_1177|M.thermoresistible__bu AYADEDGIVVXXGGNGGRGGSVILVVDPQVHTLLDFHFHPHVVAPSGKPG
Mflv_1124|M.gilvum_PYR-GCK AYSDDDGIVVVTAD------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 AYSDDDGIVVIAAG------------------------------------
MAB_0107c|M.abscessus_ATCC_199 CYADHDGVVVVPA-------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 VVGDADGVVSLPAEQVPEILERADARIRKEEQVISALRQGATTIDLYQFD
.* **::
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu QGSNRDGAAGADLEVKVPDGTVVLDEHGNLLADLTGAGTRFVAAEGGRGG
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 PTGLGN--------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu LGNAALASRARKAPGFALLGEPGQARDLVLELKTVADVGLIGYPSAGKSS
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu LVSTISAAKPKIADYPFTTLTPNLGVVSAGDETFTVADVPGLIPGASEGR
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu GLGLDFLRHIERCAVLVHVVDCATLELGRDPVSDIETLENELAAYTPTLQ
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu GDSELDDLTERPRAVVLNKIDVPEARELAEFVRDEIVQRFGWPVFLVSTA
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu TREGLRPLVFALWEMMKQYRASQPEVAPRRPVIRPVPVDETGFTVHPDGE
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu GGFVVRGVRPERWVTQTDFNNDEAVGYLADRLARLGVEEELLQLGAEPGC
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------------------------------------------------
MUL_5025|M.ulcerans_Agy99 --------------------------------------------------
Mb3883|M.bovis_AF2122/97 --------------------------------------------------
Rv3853|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00066|M.leprae_Br4923 --------------------------------------------------
MAV_0174|M.avium_104 --------------------------------------------------
TH_1177|M.thermoresistible__bu EVTIGDMTFEWEPQTPAGVDDVPLSGRGTDARLEQSERVTAAERKAARRA
Mflv_1124|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5682|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0107c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0706|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5403|M.marinum_M --------
MUL_5025|M.ulcerans_Agy99 --------
Mb3883|M.bovis_AF2122/97 --------
Rv3853|M.tuberculosis_H37Rv --------
MLBr_00066|M.leprae_Br4923 --------
MAV_0174|M.avium_104 --------
TH_1177|M.thermoresistible__bu RREPGGEP
Mflv_1124|M.gilvum_PYR-GCK --------
Mvan_5682|M.vanbaalenii_PYR-1 --------
MAB_0107c|M.abscessus_ATCC_199 --------
MSMEG_0706|M.smegmatis_MC2_155 --------