For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLDEPTRAFLERATATPPLPPGSLPLKEFRAAVESLRPSTFDRIELAEVRDLTCPGPGATTVSARFYRPE AGTSAPLVVWAHGGQWVRTTVDLLDTFFRHVAHRSGCAVLAVDYEHAPESRFPAQIEQIYAAALWAQEQN GQLDCDTGRLAIAGDSSGANLAAAVALLARERGRLQFQHQLLLVPLLDTLFDSASWRELGQDYLLSVEQL TWALRNYAPGVDRRHPLLSPLHAEDLSALPPTTIIVGEFDPLRDDGLAYARRLEADGVAVKTIDVESLLH HAMVIPKALPQGRVAVEAAADALANCW*
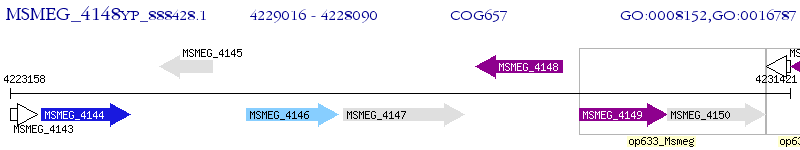
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4148 | - | - | 100% (308) | lipase |
| M. smegmatis MC2 155 | MSMEG_2888 | - | 2e-42 | 38.35% (279) | lipase |
| M. smegmatis MC2 155 | MSMEG_3829 | - | 3e-37 | 33.22% (304) | esterase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2994c | lipN | 4e-39 | 40.42% (240) | lipase/esterase LipN |
| M. gilvum PYR-GCK | Mflv_4552 | - | 1e-41 | 36.82% (258) | alpha/beta hydrolase domain-containing protein |
| M. tuberculosis H37Rv | Rv2970c | lipN | 4e-39 | 40.42% (240) | lipase/esterase LipN |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2814 | - | 8e-37 | 34.69% (294) | lipase LipH |
| M. marinum M | MMAR_0576 | - | 1e-41 | 35.40% (291) | monooxygenase |
| M. avium 104 | MAV_3025 | - | 8e-47 | 38.11% (307) | esterase |
| M. thermoresistible (build 8) | TH_2802 | - | 6e-38 | 36.24% (287) | PUTATIVE esterase |
| M. ulcerans Agy99 | MUL_1989 | lipN | 2e-39 | 40.00% (235) | lipase/esterase LipN |
| M. vanbaalenii PYR-1 | Mvan_2888 | - | 2e-47 | 40.14% (279) | alpha/beta hydrolase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M MSDAHHTDVDVVVAGAGFAGLYAAYRLRRAGHSMRIFEAGDDIGGTWYWN
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M RYPGARVDIPSVDYMYSFDPDWSRDWQWSEKYATQPEILRYLNHVADKHD
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M LRRDIQFGTRVTHAHWDDDAGRYRVRTDRGDEIICRYLIMATGCLSVPKD
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M LDIDGIENFTGELYFTSRWPHEPVDFTGKRVGVVGTGSSGIQSIPLIAAQ
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M AAQVVVFQRTPCFSIPAHNGPISPDKLAQLADDDAYRRAARYSFGGVPME
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M RSLTPTFSVSAEERRQRYERAWQLGELLEALNLYADEMINPLANSEFAEF
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M IRDKIRATVHDPQTAEALCPSGYPVGSKRLCLDSNYYATFNLPQVRLVDL
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M RNQPLLGVTKTGIDTVGESFEFDAIVFATGFDAVTGALAAIDIVGRNGLA
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M LGDKWAPGPSTYLGLTTEGFPNLFLIAGPNSPSVLSNMAVSIEQHVDFVA
Mb2994c|M.bovis_AF2122/97 --------------------------------------------------
Rv2970c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_1989|M.ulcerans_Agy99 --------------------------------------------------
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3025|M.avium_104 --------------------------------------------------
Mvan_2888|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0576|M.marinum_M DALSYLREHGFDRIEPTELAEAGWMQHSDDAASITLFPQANSWYVGANIP
Mb2994c|M.bovis_AF2122/97 -------------------------------------MTKSLPGVADLRL
Rv2970c|M.tuberculosis_H37Rv -------------------------------------MTKSLPGVADLRL
MUL_1989|M.ulcerans_Agy99 -------------------------------------MTNSLPGETDLHP
MAB_2814|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2802|M.thermoresistible__bu --------------------------------------------------
MSMEG_4148|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4552|M.gilvum_PYR-GCK ----------------------------------------------MRLD
MAV_3025|M.avium_104 ----------------------------------------------MSLE
Mvan_2888|M.vanbaalenii_PYR-1 ----------------------------------------------MSLD
MMAR_0576|M.marinum_M GKPRTFMAYTAGVDVYRVACEEVVARDYLGFRLSGPNGSQCNDGVVRRLQ
Mb2994c|M.bovis_AF2122/97 GANHPRM-WTRRVQGTVVNVGVKVLPWIPTPAKRILSAGRSVIIDGNTLD
Rv2970c|M.tuberculosis_H37Rv GANHPRM-WTRRVQGTVVNVGVKVLPWIPTPAKRILSAGRSVIIDGNTLD
MUL_1989|M.ulcerans_Agy99 ETVHATMSWLSRVQNTVTVVGAKVIPWVPDVAKRLITRGRSVIIDGNTLD
MAB_2814|M.abscessus_ATCC_1997 ---------------------------------MTATAG---------ID
TH_2802|M.thermoresistible__bu --------------------------------------------MNHPVP
MSMEG_4148|M.smegmatis_MC2_155 ----------------------------------------------MTLD
:
Mflv_4552|M.gilvum_PYR-GCK PQIAALIDALDG-GFPAVHTMTG-AQARATIRSRFTPAA-EPEPVHSVHD
MAV_3025|M.avium_104 PQIAAIIEQLDS-GFPPVQQMSG-AEARALIRSRLVPPA-QPEPVAEVAD
Mvan_2888|M.vanbaalenii_PYR-1 PQIATIIETLDS-GFPAVHTMTG-EQARAAIRSRFVPAA-EPEPVASVED
MMAR_0576|M.marinum_M PDIQMVLEQLEALGLPPLESLPA-DQARALMNEMNLARP-PGPDVGEIVD
Mb2994c|M.bovis_AF2122/97 PTLQLMLSTSRIFGVDGLAVDDDIVASRAHMRAICEAMPGPQIHVD-VTD
Rv2970c|M.tuberculosis_H37Rv PTLQLMLSTSRIFGVDGLAVDDDIVASRAHMRAICEAMPGPQIHVD-VTD
MUL_1989|M.ulcerans_Agy99 PALQLMLSGMRVVGLDGLVIDDDLAASRAHMREAMLGFPGPQIHVD-VEE
MAB_2814|M.abscessus_ATCC_1997 PIIVKILEAVPIQ----LVSPDGPQASRDAYRAIATARP-PWIEVANIED
TH_2802|M.thermoresistible__bu PLDPDAAARIASLGPPAPMRRRGLQAVRDAIESAPLPTD--MPAMASIVD
MSMEG_4148|M.smegmatis_MC2_155 EPTRAFLERATA--TPPLPPGSLPLKEFRAAVESLRPSTFDRIELAEVRD
: : :
Mflv_4552|M.gilvum_PYR-GCK MAVPVD-DGEVPVRVYRPAGP--APLPALVFAHGGGFVFCDLDSHDGLCR
MAV_3025|M.avium_104 RLVEGP-GGPLRIRVYRPEAA--APLPALVYAHGGGFVFCDLDSHDGLCR
Mvan_2888|M.vanbaalenii_PYR-1 RTVPGP-DGPVTVRIYRPAAATDGPVPTMVYAHGGGWVFCDLDSHDGLCR
MMAR_0576|M.marinum_M GTLPGA-AGPLAYRLYRPASP--GPHPVVVYFHGGGWVLGDHTSDDPLCR
Mb2994c|M.bovis_AF2122/97 LSIPGP-AGEIPARHYRPSGG--GATPLLVFYHGGGWTLGDLDTHDALCR
Rv2970c|M.tuberculosis_H37Rv LSIPGP-AGEIPARHYRPSGG--GATPLLVFYHGGGWTLGDLDTHDALCR
MUL_1989|M.ulcerans_Agy99 LTLPGP-AGDISARHYRPPGD--AAAPLLVFYHGGGWALGDLDTHDAVCR
MAB_2814|M.abscessus_ATCC_1997 RTIPGP-AGEIPVRIYSPAEG--GVRPVFVLIHGGGWVIGDLDTHDAVAR
TH_2802|M.thermoresistible__bu RAVPGP-GGDIPVRIHRPERGCGDPAPAVLYFHGGGLVMGSNHSFEPLAR
MSMEG_4148|M.smegmatis_MC2_155 LTCPGPGATTVSARFYRPEAG--TSAPLVVWAHGGQWVRTTVDLLDTFFR
: * : * * .: *** . : . *
Mflv_4552|M.gilvum_PYR-GCK NLSNRLGAVVISVGYRLAPEH-RWPTAAEDMYAVTRWVSGDAD-ALGVDP
MAV_3025|M.avium_104 NLANLVPAVVVSVDYRLAPEN-AWPAAAEDVYAATCWARDHAD-ALGADP
Mvan_2888|M.vanbaalenii_PYR-1 AFANGMSAVVVSVHYRRASEEGRWPAAAEDVYAVAAWAAETIG-ELGGDL
MMAR_0576|M.marinum_M DLCVLSDTLIVSVDYRHAPEH-RFPAALDDGWAAVQWIAEHAG-ELGGIP
Mb2994c|M.bovis_AF2122/97 LTCRDADIQVLSIDYRLAPEH-PAPAAVEDAYAAFVWAHEHASDEFGALP
Rv2970c|M.tuberculosis_H37Rv LTCRDADIQVLSIDYRLAPEH-PAPAAVEDAYAAFVWAHEHASDEFGALP
MUL_1989|M.ulcerans_Agy99 LTCRDAGIHVLAIDYRLSPEH-RAPAAIDDAFAAFEWAHAHAA-ELGALP
MAB_2814|M.abscessus_ATCC_1997 AAAVQADAVVVSVDYRLAPEH-PYPAAVEDCWAALQWVHEHAA-EIGGDP
TH_2802|M.thermoresistible__bu GLAAASGAVVVAVDYRLAPEH-PPPAQFDDAWATTRWVAAHAD-ELGLDT
MSMEG_4148|M.smegmatis_MC2_155 HVAHRSGCAVLAVDYEHAPES-RFPAQIEQIYAAALWAQEQNG-QLDCDT
. :::: *. :.* *: :: :*. * :.
Mflv_4552|M.gilvum_PYR-GCK ARIAVGGDSAGGNLAAVTALMARD----RGGPALRAQLLLYPVIAADFDT
MAV_3025|M.avium_104 ARLVVGGDSAGGNLAAVTTVMCRD----RGGPAPAAQLLIYPVIAADFDT
Mvan_2888|M.vanbaalenii_PYR-1 SALLVGGDSAGGNLAAVTALMARD----RRGPALAGQLLLYPVIAANFDT
MMAR_0576|M.marinum_M GQLVVSGWSAGAGIAAVVCHLARD----AGAPSIVGQALLTPVTDFDPTR
Mb2994c|M.bovis_AF2122/97 GRVAVGGDSAGGNLSAVVCQLARDKARYEGGPTPVLQWLLYPRTDFTAQT
Rv2970c|M.tuberculosis_H37Rv GRVAVGGDSAGGNLSAVVCQLARDKARYEGGPTPVLQWLLYPRTDFTAQT
MUL_1989|M.ulcerans_Agy99 GRVAVGGDSAGGNLAAVVSQLARD----SGGPAPIFQWLIYPRTDFAGRT
MAB_2814|M.abscessus_ATCC_1997 GRLAVGGDSAGGNLSAVMAQLARD-----NGIDLAFQVLWYPATTFDTTL
TH_2802|M.thermoresistible__bu SRLAVAGDSAGGALAAAVALAARD----RGGPELLAQLLMYPGLDRDLAA
MSMEG_4148|M.smegmatis_MC2_155 GRLAIAGDSSGANLAAAVALLARE----RGRLQFQHQLLLVPLLDTLFDS
. : :.* *:*. ::*. .*: * * *
Mflv_4552|M.gilvum_PYR-GCK ESYRLFGHGFYNPEPALRWYWDQYVPALSDR-QHPYASPLHG-ELTGLPP
MAV_3025|M.avium_104 ESYRLFGQGYYNPAPALRWYWDCYVPSTRDR-AHPYATPLNA-DLRGLPP
Mvan_2888|M.vanbaalenii_PYR-1 ESYRRFGEGFYNPLAALQWYWDQYVPNLADR-VNPYASPLHADDLSGLPP
MMAR_0576|M.marinum_M GSYLENADGYGLTAPLMQWFFDHYA-DPDVR-TDPRIAPLRAPDLSALPP
Mb2994c|M.bovis_AF2122/97 RSMGLFGNGFLLTKRDIDWFHTQYLRDSDVDPADPRLSPLLAESLSGLAP
Rv2970c|M.tuberculosis_H37Rv RSMGLFGNGFLLTKRDIDWFHTQYLRDSDVDPADPRLSPLLAESLSGLAP
MUL_1989|M.ulcerans_Agy99 RSASLFARGFLLTKRDIDWFHSQYLKGSGIEPTDPRVSPLRAESLAGLAP
MAB_2814|M.abscessus_ATCC_1997 PSMVENAEAPVLDFKTVGALSKWYLGDTNPATAGPTLVPARAENFEGLAP
TH_2802|M.thermoresistible__bu PSIVAMPEAPMLSRDDIVYLHELADLGAGTP-HDPYRVPAYATDLGGLPQ
MSMEG_4148|M.smegmatis_MC2_155 ASWRELGQDYLLSVEQLTWALRNYAPGVDRR--HPLLSPLHAEDLSALPP
* : * * . .: .*.
Mflv_4552|M.gilvum_PYR-GCK AVMVMTGHDPLRDEAVAYAQALTDAGVPVVRCEFDGAVHGFMTMPMLDIA
MAV_3025|M.avium_104 AVVVVAGHDPLRDEGLAFGAALEAAGVPTVQLRYEGGIHGFMTMPMLDIA
Mvan_2888|M.vanbaalenii_PYR-1 AITVVAGHDPLRDEGLAYTEALEAAGVETICRYFDGGVHGFMTMPTFDIC
MMAR_0576|M.marinum_M AIVVAAEFDPLRDEGIEYAEALAAAGVPTELVRARGHTHLSLTMVDVVVS
Mb2994c|M.bovis_AF2122/97 ALIAVAGFDPLRDEGESYAKALRAAGTAVDLRYLGSLTHGFLNLFQLGGG
Rv2970c|M.tuberculosis_H37Rv ALIAVAGFDPLRDEGESYAKALRAAGTAVDLRYLGSLTHGFLNLFQLGGG
MUL_1989|M.ulcerans_Agy99 ALIAVAGFDPLRDEGENYATALRAAGTPVDLRAMGSLTHGFLNLFPLGGG
MAB_2814|M.abscessus_ATCC_1997 AFIGTAQFDPLRDDGAKYAELLRDAGVPVQLHNAPTLIHGYLGYADIVPS
TH_2802|M.thermoresistible__bu TIIVTGACDPIRDWGERYAGRLRDAGVQTTVTRYPGMYHGFLMRYESTAR
MSMEG_4148|M.smegmatis_MC2_155 TTIIVGEFDPLRDDGLAYARRLEADGVAVKTIDVESLLHHAMVIPKALPQ
: **:** . : * *. . * :
Mflv_4552|M.gilvum_PYR-GCK HE-ARRRSCQELTALLHG------
MAV_3025|M.avium_104 HR-ARNEAAAALADLLRR------
Mvan_2888|M.vanbaalenii_PYR-1 GR-ARARVCADIAELVGRPVTN--
MMAR_0576|M.marinum_M GAPIRAQLAGALRGFFAIGAAVPA
Mb2994c|M.bovis_AF2122/97 SAAGTNELISALRAHLSRV-----
Rv2970c|M.tuberculosis_H37Rv SAAGTNELISALRAHLSRV-----
MUL_1989|M.ulcerans_Agy99 CAAATSELISALRAHLTRV-----
MAB_2814|M.abscessus_ATCC_1997 ATEERDLSLAALKQALHG------
TH_2802|M.thermoresistible__bu GRLALAEIGALLRAKFANPLPF--
MSMEG_4148|M.smegmatis_MC2_155 GRVAVEAAADALANCW--------
: