For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STENHEFTTVGVLGLGTMGAGITQVFAASGRDVVVLEADQDRIDAGLASISAFLDTGVAKGKLSETDKSG LLARITATTDVTDLADVDLVVESVTENAEVKKDLLGRVAAVVGVNTPICTNTSALSVTELAAALPNPSRV AGLHFFNPAPLQRTVEVVRALQTGEELVDRLVALVDTLGNKDPIVVKDRPGFLLNALLLPYLNDVISEYD DGLATAEDIDLALKLGLGYKSGPLELLDMIGLDVQLHATEAAYAATADPRYAPPPLLRQMVAAGRLGNKA GNGFRTTDSENS*
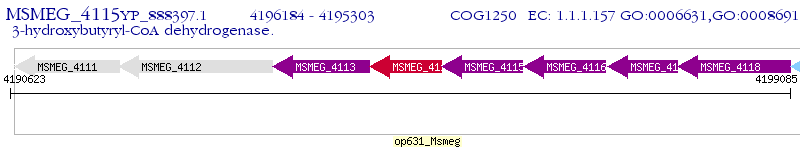
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4115 | - | - | 100% (293) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4116 | - | 4e-54 | 38.93% (280) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0912 | - | 5e-54 | 39.57% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0477 | fadB2 | 1e-55 | 41.37% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0106 | - | 8e-61 | 45.29% (276) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv0468 | fadB2 | 1e-55 | 41.37% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. leprae Br4923 | MLBr_02461 | fadB2 | 3e-54 | 39.93% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0904 | - | 1e-58 | 45.32% (278) | putative 3-hydroxyacyl-CoA dehydrogenase |
| M. marinum M | MMAR_0793 | fadB2 | 5e-56 | 41.73% (278) | 3-hydroxybutyryl-CoA dehydrogenase FadB2 |
| M. avium 104 | MAV_4681 | - | 6e-55 | 41.37% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3443 | - | 1e-46 | 38.74% (253) | PUTATIVE 3-hydroxybutyryl-CoA dehydrogenase FadB2 |
| M. ulcerans Agy99 | MUL_4537 | fadB2 | 8e-56 | 41.37% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0802 | - | 2e-60 | 44.60% (278) | 3-hydroxybutyryl-CoA dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0793|M.marinum_M -----MG----SDAAIQRVG-VVGAGQMGSGIAEVSVRAGVEVTVFETTE
MUL_4537|M.ulcerans_Agy99 -----MG----SDAAIQRVG-VVGAGQMGSGIAEVSVRAGVEVTAFETTE
MAV_4681|M.avium_104 MHDKTIGGETVSDAAIQRVG-VVGAGQMGSGIAEVSVRADVDVTVFETTE
MLBr_02461|M.leprae_Br4923 ----------MSDAAIQRVG-VVGAGQMGSGIAEVAVRAGVGVTVFETTD
Mb0477|M.bovis_AF2122/97 -----------MSDAIQRVG-VVGAGQMGSGIAEVSARAGVEVTVFEPAE
Rv0468|M.tuberculosis_H37Rv -----------MSDAIQRVG-VVGAGQMGSGIAEVSARAGVEVTVFEPAE
TH_3443|M.thermoresistible__bu -------------------------------LRWWHARERCPX-VYDISE
Mflv_0106|M.gilvum_PYR-GCK -------------MSIERVG-VVGAGQMGSGIAEVAAKAGANVVVYEPSD
Mvan_0802|M.vanbaalenii_PYR-1 -------------MSIERVG-VVGAGQMGSGIAEVAAKAGANVVVYEPSE
MSMEG_4115|M.smegmatis_MC2_155 --------MSTENHEFTTVG-VLGLGTMGAGITQVFAASGRDVVVLEADQ
MAB_0904|M.abscessus_ATCC_1997 --------MTATPHSVPTVVTVIGGGPMGAGIAQVFAAAGANVTIVETGR
: . :
MMAR_0793|M.marinum_M ALITAGRNRIVKSLERGVSSGKVTERERDRALSRLTFTTDLNDL-ADRQL
MUL_4537|M.ulcerans_Agy99 ALITAGRNRIVKSLERGVSSGKVTERERDRALSRLTFTTDLNDL-ADRQL
MAV_4681|M.avium_104 ALITAGRNRIVKSLERGVSAGKVTERERDRALSKLTFTTDLKDL-ADRQL
MLBr_02461|M.leprae_Br4923 TLVTAGRNRIVKSLERGVSAGKVTERERDSALGHLVFTTDLNDL-ADRQL
Mb0477|M.bovis_AF2122/97 ALITAGRNRIVKSLERAVSAGKVTERERDRALGLLTFTTDLNDL-SDRQL
Rv0468|M.tuberculosis_H37Rv ALITAGRNRIVKSLERAVSAGKVTERERDRALGLLTFTTDLNDL-SDRQL
TH_3443|M.thermoresistible__bu ELTTAGRERIAKSLTRGVQAGKLSEDERSAALDRLRFTTTLADL-SDRQL
Mflv_0106|M.gilvum_PYR-GCK ALVNAGRERITASLERAKAKGKLAADDFEITLARLTFTTDLADL-ADRQL
Mvan_0802|M.vanbaalenii_PYR-1 ALVAAGRDRITASLERAKAKGKLAADDFDATVARLTFTTNLGDL-ADRQL
MSMEG_4115|M.smegmatis_MC2_155 DRIDAGLASISAFLDTGVAKGKLSETDKSGLLARITATTDVTDL-ADVDL
MAB_0904|M.abscessus_ATCC_1997 DRADAALQRVSAGLDRAAAKNRLAEAPEH-VLSRVDVVCSLTELPTDSEL
*. : * . .::: : : . : :* :* :*
MMAR_0793|M.marinum_M VIEAIIEDEAVKTKVFADLDRVVTDPDAVLASNTSSIPIMKVAAATKNPR
MUL_4537|M.ulcerans_Agy99 VIEAIIEDEAVKTKVFADLDRVVTDPDAVLASNTSSIPIMKVAAATKNPR
MAV_4681|M.avium_104 VIEAIIEDDAVKAQVFAELDRVVTDPDAVLASNTSSIPIMKLAAATKNPQ
MLBr_02461|M.leprae_Br4923 VIEAIVEDEAVKTKVFAELDRVITDPDAVLASNTSSIPIMKIAAATKNPQ
Mb0477|M.bovis_AF2122/97 VIEAVVEDEAVKSEIFAELDRVVTDPDAVLASNTSSIPIMKVAAATKQPQ
Rv0468|M.tuberculosis_H37Rv VIEAVVEDEAVKSEIFAELDRVVTDPDAVLASNTSSIPIMKVAAATKQPQ
TH_3443|M.thermoresistible__bu VIEAIVEDENVKATVFAELDELLTDPDAVLASNTSSIPIMKLAAATKNPG
Mflv_0106|M.gilvum_PYR-GCK VIEAIIEDEAVKAKVFAQLDEVVTDPDAVLASNTSSIPIMKIAAATKNPS
Mvan_0802|M.vanbaalenii_PYR-1 VIEAIVENEAVKAKVFAELDAVVTDPDAVLASNTSSIPIMKIAAATKNPS
MSMEG_4115|M.smegmatis_MC2_155 VVESVTENAEVKKDLLGRVAAVV-GVNTPICTNTSALSVTELAAALPNPS
MAB_0904|M.abscessus_ATCC_1997 VVEAVPENINLKIEVLGAAEKLV-EPTTVLASNTSSLSITELAAALLRPE
*:*:: *: :* ::. :: : :.:***::.: ::*** .*
MMAR_0793|M.marinum_M RVLGLHFFNPVPVLPLVELVSTLVTDEAAVARTEEFASAVLGKQVVRCSD
MUL_4537|M.ulcerans_Agy99 RVLGLHFFNPVPVLPLVELVNTLVTDEAAVARTEEFASAVLGKQVVRCSD
MAV_4681|M.avium_104 RVLGLHFFNPVPVLPLVELVSTLVTDEAAAARTEEFASAVLGKQVVRCSD
MLBr_02461|M.leprae_Br4923 RVLGLHFFNPVPVLPLVELVSTLVTDEAAATRAEEFASGVLGKQVVRCSD
Mb0477|M.bovis_AF2122/97 RVLGLHFFNPVPVLPLVELVRTLVTDEAAAARTEEFASTVLGKQVVRCSD
Rv0468|M.tuberculosis_H37Rv RVLGLHFFNPVPVLPLVELVRTLVTDEAAAARTEEFASTVLGKQVVRCSD
TH_3443|M.thermoresistible__bu RVLGLHFFNPVPVLPLVELINTLVTTDEAIARTEKFAAEVLGKQVVRCGD
Mflv_0106|M.gilvum_PYR-GCK RVLGLHFFNPVPVLPLVEVINTLVTSEEAVARVEQFASEVLGKKVVRCGD
Mvan_0802|M.vanbaalenii_PYR-1 RVLGLHFFNPVPVLPLVELVNTLVTSDDAIARVERFAGEVLGKKVVRCGD
MSMEG_4115|M.smegmatis_MC2_155 RVAGLHFFNPAPLQRTVEVVRALQTGEELVDRLVALVDTLGNKDPIVVKD
MAB_0904|M.abscessus_ATCC_1997 RVVGMHFFNPVPTSQLVEIVRAPETSDDTVAAALAWVRALG-KADVVARD
** *:*****.* **:: : * : . : * : *
MMAR_0793|M.marinum_M RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSD
MUL_4537|M.ulcerans_Agy99 RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSD
MAV_4681|M.avium_104 RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSD
MLBr_02461|M.leprae_Br4923 RSGFVVNALLVPYLLSAIRIVEAGFATVEDVDKAVLAGLSHPMGPLRLSD
Mb0477|M.bovis_AF2122/97 RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSD
Rv0468|M.tuberculosis_H37Rv RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSD
TH_3443|M.thermoresistible__bu RSGFVVNALLVPYLLSAIRMVESGYASVEDVDKAVVAGLSHPMGPLRLSD
Mflv_0106|M.gilvum_PYR-GCK RSGFIVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMGPLRLSD
Mvan_0802|M.vanbaalenii_PYR-1 RSGFVVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMGPLRLSD
MSMEG_4115|M.smegmatis_MC2_155 RPGFLLNALLLPYLNDVISEYDDGLATAEDIDLALKLGLGYKSGPLELLD
MAB_0904|M.abscessus_ATCC_1997 SPGFASSRLGVALGLEAIRMLEEGVADAESIDRAMELGYRHPMGPLRSTD
.** . * :. ..* : * * .*.:* *: * : ***. *
MMAR_0793|M.marinum_M LVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGQGFYTY-
MUL_4537|M.ulcerans_Agy99 LVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGQGFYTY-
MAV_4681|M.avium_104 LVGLDTLKLIADKMFEEFKEPQYGAPPLLLRMVEAGRLGKKTGQGFYTY-
MLBr_02461|M.leprae_Br4923 LVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGLLGKKSGQGFYTY-
Mb0477|M.bovis_AF2122/97 LVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGRGFYTY-
Rv0468|M.tuberculosis_H37Rv LVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGRGFYTY-
TH_3443|M.thermoresistible__bu LVGLDTLKLIADKMFDEFKEPHYGPPPLLVRMVEAGHLGKKTGRGFYTY-
Mflv_0106|M.gilvum_PYR-GCK LIGLDTMKLIADSMYDEYKDPNFAPPPLLQRMVEAGQLGKKSGKGFYSY-
Mvan_0802|M.vanbaalenii_PYR-1 LIGLDTMKLIADSMYDEYKDPHYAPPPLLQRMVEAGQLGKKSGKGFYTY-
MSMEG_4115|M.smegmatis_MC2_155 MIGLDVQLHATEAAYAATADPRYAPPPLLRQMVAAGRLGNKAGNGFRTTD
MAB_0904|M.abscessus_ATCC_1997 LVGLDVRLAIAEH-LCATLGPRFTPPKLLRAKVAEGNLGRKTGQGFFRWS
::***. :: *.: .* ** * * **.*:*.**
MMAR_0793|M.marinum_M ----
MUL_4537|M.ulcerans_Agy99 ----
MAV_4681|M.avium_104 ----
MLBr_02461|M.leprae_Br4923 ----
Mb0477|M.bovis_AF2122/97 ----
Rv0468|M.tuberculosis_H37Rv ----
TH_3443|M.thermoresistible__bu ----
Mflv_0106|M.gilvum_PYR-GCK ----
Mvan_0802|M.vanbaalenii_PYR-1 ----
MSMEG_4115|M.smegmatis_MC2_155 SENS
MAB_0904|M.abscessus_ATCC_1997 S---