For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKISTADRKTVAVLGAGTMGSGIATVMARAGHRTILYDINEANLERGIDTVHGFFDKSVRLGKLDATAGQ AAKDSLSGSTELKDLAPCDVVVEAVFEDLSLKKETFGRLDDIVPPTTLFHTNTSTLSVTGIASGSRLRER VVGTHYCNPAPLMKLVEVANGRHTADWAHKATLEFLASLGKTSVVTKDRPGFIVNRFLIPWENSCIEALE AGVATKEEIDTAVLGALGHPMGPFRLLDIVGMDIHQQVATRLYEQLRDPKFFPPPMVERMVAAGDLGRKT GRGFYEYDDTRLFGS*
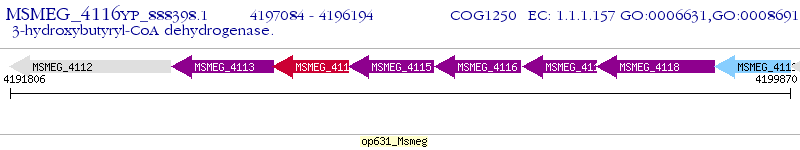
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4116 | - | - | 100% (296) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0912 | - | 4e-56 | 40.86% (279) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4115 | - | 4e-54 | 38.93% (280) | 3-hydroxybutyryl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0477 | fadB2 | 4e-57 | 41.22% (279) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0106 | - | 5e-60 | 42.29% (279) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv0468 | fadB2 | 4e-57 | 41.22% (279) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. leprae Br4923 | MLBr_02461 | fadB2 | 3e-55 | 39.02% (287) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4094c | - | 5e-55 | 40.86% (279) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. marinum M | MMAR_0793 | fadB2 | 5e-56 | 39.86% (286) | 3-hydroxybutyryl-CoA dehydrogenase FadB2 |
| M. avium 104 | MAV_4681 | - | 3e-56 | 39.72% (287) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3443 | - | 6e-50 | 39.76% (254) | PUTATIVE 3-hydroxybutyryl-CoA dehydrogenase FadB2 |
| M. ulcerans Agy99 | MUL_4537 | fadB2 | 2e-56 | 40.21% (286) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0802 | - | 2e-58 | 42.29% (279) | 3-hydroxybutyryl-CoA dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0793|M.marinum_M -----MG----SDAAIQRVGVVGAGQMGSGIAEVSVRAGVEVTVFETTEA
MUL_4537|M.ulcerans_Agy99 -----MG----SDAAIQRVGVVGAGQMGSGIAEVSVRAGVEVTAFETTEA
MAV_4681|M.avium_104 MHDKTIGGETVSDAAIQRVGVVGAGQMGSGIAEVSVRADVDVTVFETTEA
MLBr_02461|M.leprae_Br4923 ----------MSDAAIQRVGVVGAGQMGSGIAEVAVRAGVGVTVFETTDT
Mb0477|M.bovis_AF2122/97 -----------MSDAIQRVGVVGAGQMGSGIAEVSARAGVEVTVFEPAEA
Rv0468|M.tuberculosis_H37Rv -----------MSDAIQRVGVVGAGQMGSGIAEVSARAGVEVTVFEPAEA
MAB_4094c|M.abscessus_ATCC_199 -----------MTDGITRVGVIGAGQMGAGIAEVSARAGVDVLVFETTEA
TH_3443|M.thermoresistible__bu ------------------------------LRWWHARERCPX-VYDISEE
Mflv_0106|M.gilvum_PYR-GCK -------------MSIERVGVVGAGQMGSGIAEVAAKAGANVVVYEPSDA
Mvan_0802|M.vanbaalenii_PYR-1 -------------MSIERVGVVGAGQMGSGIAEVAAKAGANVVVYEPSEA
MSMEG_4116|M.smegmatis_MC2_155 -------MSKISTADRKTVAVLGAGTMGSGIATVMARAGHRTILYDINEA
: .: :: :
MMAR_0793|M.marinum_M LITAGRNRIVKSLERGVSSGKVTERERDRALSRLTFTTDLNDLADRQLVI
MUL_4537|M.ulcerans_Agy99 LITAGRNRIVKSLERGVSSGKVTERERDRALSRLTFTTDLNDLADRQLVI
MAV_4681|M.avium_104 LITAGRNRIVKSLERGVSAGKVTERERDRALSKLTFTTDLKDLADRQLVI
MLBr_02461|M.leprae_Br4923 LVTAGRNRIVKSLERGVSAGKVTERERDSALGHLVFTTDLNDLADRQLVI
Mb0477|M.bovis_AF2122/97 LITAGRNRIVKSLERAVSAGKVTERERDRALGLLTFTTDLNDLSDRQLVI
Rv0468|M.tuberculosis_H37Rv LITAGRNRIVKSLERAVSAGKVTERERDRALGLLTFTTDLNDLSDRQLVI
MAB_4094c|M.abscessus_ATCC_199 LTTAGRDRITKSLDRGVSAGKITERERDAAVANLKFTTDLADFADRQLVI
TH_3443|M.thermoresistible__bu LTTAGRERIAKSLTRGVQAGKLSEDERSAALDRLRFTTTLADLSDRQLVI
Mflv_0106|M.gilvum_PYR-GCK LVNAGRERITASLERAKAKGKLAADDFEITLARLTFTTDLADLADRQLVI
Mvan_0802|M.vanbaalenii_PYR-1 LVAAGRDRITASLERAKAKGKLAADDFDATVARLTFTTNLGDLADRQLVI
MSMEG_4116|M.smegmatis_MC2_155 NLERGIDTVHGFFDKSVRLGKLDATAGQAAKDSLSGSTELKDLAPCDVVV
* : : : :. **: . : * :* * *:: ::*:
MMAR_0793|M.marinum_M EAIIEDEAVKTKVFADLDRVVTDPDAVLASNTSSIPIMKVAAATKNPRRV
MUL_4537|M.ulcerans_Agy99 EAIIEDEAVKTKVFADLDRVVTDPDAVLASNTSSIPIMKVAAATKNPRRV
MAV_4681|M.avium_104 EAIIEDDAVKAQVFAELDRVVTDPDAVLASNTSSIPIMKLAAATKNPQRV
MLBr_02461|M.leprae_Br4923 EAIVEDEAVKTKVFAELDRVITDPDAVLASNTSSIPIMKIAAATKNPQRV
Mb0477|M.bovis_AF2122/97 EAVVEDEAVKSEIFAELDRVVTDPDAVLASNTSSIPIMKVAAATKQPQRV
Rv0468|M.tuberculosis_H37Rv EAVVEDEAVKSEIFAELDRVVTDPDAVLASNTSSIPIMKVAAATKQPQRV
MAB_4094c|M.abscessus_ATCC_199 EAVIEDESIKSSIFAKLDEVITDPDAVLASNTSSIPIMKIAAATKNPGRV
TH_3443|M.thermoresistible__bu EAIVEDENVKATVFAELDELLTDPDAVLASNTSSIPIMKLAAATKNPGRV
Mflv_0106|M.gilvum_PYR-GCK EAIIEDEAVKAKVFAQLDEVVTDPDAVLASNTSSIPIMKIAAATKNPSRV
Mvan_0802|M.vanbaalenii_PYR-1 EAIVENEAVKAKVFAELDAVVTDPDAVLASNTSSIPIMKIAAATKNPSRV
MSMEG_4116|M.smegmatis_MC2_155 EAVFEDLSLKKETFGRLDDIVP-PTTLFHTNTSTLSVTGIASGSRLRERV
**:.*: :* *. ** ::. * ::: :***::.: :*:.:: **
MMAR_0793|M.marinum_M LGLHFFNPVPVLPLVELVSTLVTDEAAVARTEEFASAVLGKQVVRCSDRS
MUL_4537|M.ulcerans_Agy99 LGLHFFNPVPVLPLVELVNTLVTDEAAVARTEEFASAVLGKQVVRCSDRS
MAV_4681|M.avium_104 LGLHFFNPVPVLPLVELVSTLVTDEAAAARTEEFASAVLGKQVVRCSDRS
MLBr_02461|M.leprae_Br4923 LGLHFFNPVPVLPLVELVSTLVTDEAAATRAEEFASGVLGKQVVRCSDRS
Mb0477|M.bovis_AF2122/97 LGLHFFNPVPVLPLVELVRTLVTDEAAAARTEEFASTVLGKQVVRCSDRS
Rv0468|M.tuberculosis_H37Rv LGLHFFNPVPVLPLVELVRTLVTDEAAAARTEEFASTVLGKQVVRCSDRS
MAB_4094c|M.abscessus_ATCC_199 LGLHFFNPVPVLPLVELISTLVTTPEAAARTEAFASEVLGKQVVRAADRS
TH_3443|M.thermoresistible__bu LGLHFFNPVPVLPLVELINTLVTTDEAIARTEKFAAEVLGKQVVRCGDRS
Mflv_0106|M.gilvum_PYR-GCK LGLHFFNPVPVLPLVEVINTLVTSEEAVARVEQFASEVLGKKVVRCGDRS
Mvan_0802|M.vanbaalenii_PYR-1 LGLHFFNPVPVLPLVELVNTLVTSDDAIARVERFAGEVLGKKVVRCGDRS
MSMEG_4116|M.smegmatis_MC2_155 VGTHYCNPAPLMKLVEVANGRHTADWAHKATLEFLAS-LGKTSVVTKDRP
:* *: **.*:: ***: * * . * . *** * **.
MMAR_0793|M.marinum_M GFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSDLV
MUL_4537|M.ulcerans_Agy99 GFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSDLV
MAV_4681|M.avium_104 GFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSDLV
MLBr_02461|M.leprae_Br4923 GFVVNALLVPYLLSAIRIVEAGFATVEDVDKAVLAGLSHPMGPLRLSDLV
Mb0477|M.bovis_AF2122/97 GFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSDLV
Rv0468|M.tuberculosis_H37Rv GFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMGPLRLSDLV
MAB_4094c|M.abscessus_ATCC_199 GFVVNFLLVPYLLAAIRMAESGFATVEDIDKAVVAGLSHPMGPLRLSDLV
TH_3443|M.thermoresistible__bu GFVVNALLVPYLLSAIRMVESGYASVEDVDKAVVAGLSHPMGPLRLSDLV
Mflv_0106|M.gilvum_PYR-GCK GFIVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMGPLRLSDLI
Mvan_0802|M.vanbaalenii_PYR-1 GFVVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMGPLRLSDLI
MSMEG_4116|M.smegmatis_MC2_155 GFIVNRFLIPWENSCIEALEAGVATKEEIDTAVLGALGHPMGPFRLLDIV
**:** :*:*: :.*. *:* *: *::*.**:..*.*****:** *::
MMAR_0793|M.marinum_M GLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGQGFYTY---
MUL_4537|M.ulcerans_Agy99 GLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGQGFYTY---
MAV_4681|M.avium_104 GLDTLKLIADKMFEEFKEPQYGAPPLLLRMVEAGRLGKKTGQGFYTY---
MLBr_02461|M.leprae_Br4923 GLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGLLGKKSGQGFYTY---
Mb0477|M.bovis_AF2122/97 GLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGRGFYTY---
Rv0468|M.tuberculosis_H37Rv GLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGRGFYTY---
MAB_4094c|M.abscessus_ATCC_199 GLDTLKLIADSMYDEYKEPLYAAPPLLLRMVEAGRLGKKSGVGFYEYKAK
TH_3443|M.thermoresistible__bu GLDTLKLIADKMFDEFKEPHYGPPPLLVRMVEAGHLGKKTGRGFYTY---
Mflv_0106|M.gilvum_PYR-GCK GLDTMKLIADSMYDEYKDPNFAPPPLLQRMVEAGQLGKKSGKGFYSY---
Mvan_0802|M.vanbaalenii_PYR-1 GLDTMKLIADSMYDEYKDPHYAPPPLLQRMVEAGQLGKKSGKGFYTY---
MSMEG_4116|M.smegmatis_MC2_155 GMDIHQQVATRLYEQLRDPKFFPPPMVERMVAAGDLGRKTGRGFYEYDDT
*:* : :* :::: ::* : .**:: *** ** **:*:* *** *
MMAR_0793|M.marinum_M -----
MUL_4537|M.ulcerans_Agy99 -----
MAV_4681|M.avium_104 -----
MLBr_02461|M.leprae_Br4923 -----
Mb0477|M.bovis_AF2122/97 -----
Rv0468|M.tuberculosis_H37Rv -----
MAB_4094c|M.abscessus_ATCC_199 -----
TH_3443|M.thermoresistible__bu -----
Mflv_0106|M.gilvum_PYR-GCK -----
Mvan_0802|M.vanbaalenii_PYR-1 -----
MSMEG_4116|M.smegmatis_MC2_155 RLFGS