For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RSYVGKRLGQAVLVLWAAYTVSFLLLSALPGDAVNNRIQNPEAQMTPEQAQVLIEYYGLDRPLWEQYVTS LAAVSRGDLGYSLTDGRTVTELLGSSLPSTLALTGLALVLGVVFSATIGLAANYAPWKRLRDTVASMPAL FGSIPTFVVGILLLQFFSFGLHLIPASDNGSFVALLAPAVTLGILIAAPLAQVFTTSIRSARQQPFAHVL TARGAGEAYVFRHGVLRNSSMPVLTLLGLACGELIAGAVVTEAVFARRGIGSLTVGAVSTQDLPVLQGVV LIATLAYVTVNLVVDFAYPFIDPRVLVDGRTQQVTRARRRPTSAAANREATAVPSLRVPRRVPVEVSMP*
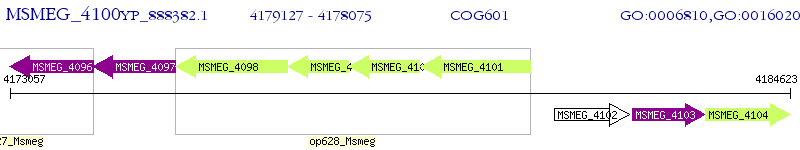
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4100 | - | - | 100% (350) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_2846 | - | 2e-60 | 39.12% (317) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_6866 | - | 6e-49 | 39.67% (300) | dipeptide transport system permease protein DppB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3689c | dppB | 2e-34 | 30.16% (305) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2760 | - | 2e-39 | 35.62% (320) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3665c | dppB | 2e-34 | 30.16% (305) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01124 | - | 2e-21 | 25.99% (327) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0427 | - | 2e-34 | 31.91% (304) | peptide ABC transporter transmembrane protein |
| M. marinum M | MMAR_5153 | dppB | 5e-33 | 30.49% (305) | dipeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_3420 | - | 6e-57 | 37.42% (318) | ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_2645 | - | 9e-43 | 36.70% (327) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4239 | dppB | 5e-33 | 30.49% (305) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3774 | - | 6e-39 | 35.29% (323) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3689c|M.bovis_AF2122/97 ----------------MGWYVARRVAVMVPVFLGATLLIYGMVFLLPGDP
Rv3665c|M.tuberculosis_H37Rv ----------------MGWYVARRVAVMVPVFLGATLLIYGMVFLLPGDP
MMAR_5153|M.marinum_M ----------------MVWYVARRILAMVPVFLGATLLIYGMVFLLPGDP
MUL_4239|M.ulcerans_Agy99 ----------------MVWYVARRILAMVPVFLGATLLIYGMVFLLPGDP
MAB_0427|M.abscessus_ATCC_1997 MTTVLGNVRQFWQSAGLFRYIAKRLLLAIPVLIGTSLLIYSLVYALPGDP
Mflv_2760|M.gilvum_PYR-GCK --------------MPILRFVARRLLYSAVVMFGVLLVVFALVHLVPGDP
Mvan_3774|M.vanbaalenii_PYR-1 ----------MSTAMPVLRFVARRLLFSAVVMIGVLVVVFALVHLVPGDP
TH_2645|M.thermoresistible__bu ---------MRFVTHPIARFLGRRLLYSLAVLFGVLVVVFGLMQLVPGDP
MLBr_01124|M.leprae_Br4923 ----------------MMRFLARRLLNYVVLLALASFLTYCLTSMAFAPL
MSMEG_4100|M.smegmatis_MC2_155 ----------------MRSYVGKRLGQAVLVLWAAYTVSFLLLSALPGDA
MAV_3420|M.avium_104 ----------------MLGYVLARIGQSAIVLLAVFSLVFWGVSILPADP
: :: *: :: . : : .
Mb3689c|M.bovis_AF2122/97 VAALA--GDRPLTPAVAAQLRSHYHLDDPFLVQYLRYLGGILHGDLGRAY
Rv3665c|M.tuberculosis_H37Rv VAALA--GDRPLTPAVAAQLRSHYHLDDPFLVQYLRYLGGILHGDLGRAY
MMAR_5153|M.marinum_M LAAIA--GDRPLTPAVAQQLRARYHLDDPFLVQYAHYLGGVLHGDLGRAY
MUL_4239|M.ulcerans_Agy99 LAAIA--GDRPLTPAVAQQLRARYHLDDPFLVQYAHYLGGVLHGDLGRAY
MAB_0427|M.abscessus_ATCC_1997 IRALA--GDRPFTPEVMAQLRERFHLNDPFLVRYGKYIVGFLHGDFGTDF
Mflv_2760|M.gilvum_PYR-GCK VRIAL--GTR-YTPEAYEALRAASGLDRPLVSQFFGYIGSALTGDLGVSF
Mvan_3774|M.vanbaalenii_PYR-1 VRIAL--GTR-YTPEAYEALRAASGLDRPIVSQFFGYLGSALTGDLGVSF
TH_2645|M.thermoresistible__bu VRIAL--GTR-YTPEAYQALRSASGLDRPILEQFVGYLTSAVRGDLGVSF
MLBr_01124|M.leprae_Br4923 DSLLQ--RSPHPPQAMVDAKAHALGLDKPIPIRYANWASHAIRGDFGTTI
MSMEG_4100|M.smegmatis_MC2_155 VNNRIQNPEAQMTPEQAQVLIEYYGLDRPLWEQYVTSLAAVSRGDLGYSL
MAV_3420|M.avium_104 AAIFVAKGEGYFNPDIVAQVKAFYGYDRPLWVQYFAQLNQVLHGHFGFSL
: *: :: *.:*
Mb3689c|M.bovis_AF2122/97 S-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGLRQG-GIFDS
Rv3665c|M.tuberculosis_H37Rv S-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGLRQG-GIFDS
MMAR_5153|M.marinum_M S-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGLRQG-GIFDA
MUL_4239|M.ulcerans_Agy99 S-GLPVSAVLAHAFPVTIRLAWIALAVEAVLGIGFGVIAGLRQG-GIFDA
MAB_0427|M.abscessus_ATCC_1997 Q-QRPVAQTVSQRLPVTLRLTVVAVAFETIIGITAGVLCAVRRG-SFYDN
Mflv_2760|M.gilvum_PYR-GCK RNGDPVTVTLLERLPATLSLGLAGIVIALAIALPAGIYAALREG-RLSDA
Mvan_3774|M.vanbaalenii_PYR-1 RNGDPVTLTLLERLPATLSLGLVGIVIALAIALPAGIYAALREG-RLSDA
TH_2645|M.thermoresistible__bu RNGDPVTLVLLERLPATVSLALAGIAIALAIALPAGIWSALRDG-RAGAA
MLBr_01124|M.leprae_Br4923 T-GQPVGSTLGRRVGVSLRLLIIGSLTGTVLGVTAGAWGAIRQY-QLSDR
MSMEG_4100|M.smegmatis_MC2_155 TDGRTVTELLGSSLPSTLALTGLALVLGVVFSATIG-LAANYAPWKRLRD
MAV_3420|M.avium_104 SSGQAVTDRIGGVIGETLKLAATATAFAVLFAVSVTALATTCAP---VRS
.* : . :: * . :.
Mb3689c|M.bovis_AF2122/97 AVLVTGLVIIAIPIFVLG---FLAQFLFGVQLEIAPVTVGERAS------
Rv3665c|M.tuberculosis_H37Rv AVLVTGLVIIAIPIFVLG---FLAQFLFGVQLEIAPVTVGERAS------
MMAR_5153|M.marinum_M TVLLTGLIIIAVPIFVLG---FLAQYLFGVRLGLAPVTVGARTT------
MUL_4239|M.ulcerans_Agy99 TVLLTGLIIIAAPIFVLG---FLAQYLFGVRLGLAPVTVGARTT------
MAB_0427|M.abscessus_ATCC_1997 LVLVTTTLLVSMPVFVLG---FLAQYLFGFKLGWFPI-AGIRDG------
Mflv_2760|M.gilvum_PYR-GCK LVRISSQFGVSIPDFWMG---ILLIALFSTVLGWLPT-SGYRPLF-----
Mvan_3774|M.vanbaalenii_PYR-1 LVRISSQFGVSIPDFWMG---ILLIALFSTVLGWLPT-SGYRPLT-----
TH_2645|M.thermoresistible__bu AVRVASQVGVSVPDFWMG---ILLIALFASTLGWLPT-SGYRALS-----
MLBr_01124|M.leprae_Br4923 VVTLLALLVLSTPTFVIANLLILSALRVNWAFGIQLFDYTGETSPGVYGG
MSMEG_4100|M.smegmatis_MC2_155 TVASMPALFGSIPTFVVG---ILLLQFFSFGLHLIPASDNGSFVA-----
MAV_3420|M.avium_104 VLRAIPPLFGAVPTFWLG---LVVLQVFSVQLGLISLFPDGSVTS-----
: . : * * :. :: . :
Mb3689c|M.bovis_AF2122/97 --------VGRLLLPGIVLGAMSFAYVVRLTRSAVAANAHADYVRTATAK
Rv3665c|M.tuberculosis_H37Rv --------VGRLLLPGIVLGAMSFAYVVRLTRSAVAANAHADYVRTATAK
MMAR_5153|M.marinum_M --------LDRLLLPGMVLGAVSFAYVVRLTRSAVAANAHADYVRTATAK
MUL_4239|M.ulcerans_Agy99 --------LDRLLLPGMVLGAVSFAYVVRLTRSAVAANAHADYVRTATAK
MAB_0427|M.abscessus_ATCC_1997 --------WYSFLLPGLVLASLSMAYVARLTRTSMLETMDAEFVRTARAK
Mflv_2760|M.gilvum_PYR-GCK --EDPAGWLRHIILPALTVAVVAAAIMTRYVRAAVLEVASMGYVRTARSK
Mvan_3774|M.vanbaalenii_PYR-1 --EDPAGWLRHVVLPGLTVGVVAAAIMTRYVRSAVLEVAAMGYVRTARSK
TH_2645|M.thermoresistible__bu --DDPVGWLRHLVLPALTVGLVAAAIMTRYIRSAVLEVAAAGHVRTARSK
MLBr_01124|M.leprae_Br4923 AWAHVIDRLRHLILPTLTLALVGAAGYSRYQRNAMLDVLSQDFIRAARAK
MSMEG_4100|M.smegmatis_MC2_155 -----------LLAPAVTLGILIAAPLAQVFTTSIRSARQQPFAHVLTAR
MAV_3420|M.avium_104 -----------LLVAALVLAVPVSAPIAQVLGKNIDATLALPHVNTARAK
.: . :.:. * : : . .. ::
Mb3689c|M.bovis_AF2122/97 GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAIVTEGIFNIHGVGGV
Rv3665c|M.tuberculosis_H37Rv GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAIVTEGIFNIHGVGGV
MMAR_5153|M.marinum_M GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAVVTEGIFNIHGVGGV
MUL_4239|M.ulcerans_Agy99 GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAVVTEGIFNIHGVGGV
MAB_0427|M.abscessus_ATCC_1997 GISESRILLRHTLRNSLIPVVTFIGADVGALLAGAVVTESVFNIPGLGRA
Mflv_2760|M.gilvum_PYR-GCK GLTPRVVTFRHTVRNALVPILTITGIQLATLLGGVIVVEVVFAWPGLGRL
Mvan_3774|M.vanbaalenii_PYR-1 GLSPRIVTFRHTVRNALVPILTITGIQLATLLGGVIVVEVVFAWPGLGRL
TH_2645|M.thermoresistible__bu GLSPWVLTVRHTVRNALIPVLTITGIQLATILGGVIVVEVVFAWPGLGRL
MLBr_01124|M.leprae_Br4923 GLTRRRALLKHGLRTALIPMATLFAYGVAGLVTGALFVEKIFGWHGMGEW
MSMEG_4100|M.smegmatis_MC2_155 GAGEAYVFRHGVLRNSSMPVLTLLGLACGELIAGAVVTEAVFARRGIGSL
MAV_3420|M.avium_104 GGTPGWVIRKHVVKNAAGPALTVTATTVGALLGGSVVTETVFSRSGVGAV
* ::.: * *. . . :: * :..* :* *:*
Mb3689c|M.bovis_AF2122/97 LYQAVTRQETPTVVSIVTVLVLIYLITNLLVDLLYAALDPRIRYG-----
Rv3665c|M.tuberculosis_H37Rv LYQAVTRQETPTVVSIVTVLVLIYLITNLLVDLLYAALDPRIRYG-----
MMAR_5153|M.marinum_M LYQAVTRQEAPTVVSIVTVLVMVYLLTNLLVDLLYAALDPRIRYG-----
MUL_4239|M.ulcerans_Agy99 LYQAVTRQEAPTVVSIVTVLVMVYLLTNLLVDLLYAALDPRIRYG-----
MAB_0427|M.abscessus_ATCC_1997 TFDAVRNQEGAVVVSILTLIVFFYIFFNLVVDILYALLDPRIRYE-----
Mflv_2760|M.gilvum_PYR-GCK VFNAVAARDYPLIQGAVLLIAALFLLINLLVDVLYAVVDPRIRLG-----
Mvan_3774|M.vanbaalenii_PYR-1 VFNAVAARDYPLIQGAVLLIAALFLVINLLVDVLYAVVDPRIRLG-----
TH_2645|M.thermoresistible__bu VYNAVAARDYPVIQGAVLLIAALFLLINLLVDLLYAIVDPRIRLA-----
MLBr_01124|M.leprae_Br4923 MVQGVATQDTNIIAAITIFFGTVILLAGLLSDVIYAALDPRVRVS-----
MSMEG_4100|M.smegmatis_MC2_155 TVGAVSTQDLPVLQGVVLIATLAYVTVNLVVDFAYPFIDPRVLVDGRTQQ
MAV_3420|M.avium_104 LLQAVSSQDISLIQGLVLLTAVAIVAANLAVDLIYPLLDPR---------
.* :: : . . : .* *. *. :***
Mb3689c|M.bovis_AF2122/97 ------------------------------------
Rv3665c|M.tuberculosis_H37Rv ------------------------------------
MMAR_5153|M.marinum_M ------------------------------------
MUL_4239|M.ulcerans_Agy99 ------------------------------------
MAB_0427|M.abscessus_ATCC_1997 ------------------------------------
Mflv_2760|M.gilvum_PYR-GCK ------------------------------------
Mvan_3774|M.vanbaalenii_PYR-1 ------------------------------------
TH_2645|M.thermoresistible__bu ------------------------------------
MLBr_01124|M.leprae_Br4923 ------------------------------------
MSMEG_4100|M.smegmatis_MC2_155 VTRARRRPTSAAANREATAVPSLRVPRRVPVEVSMP
MAV_3420|M.avium_104 VTRVQRRGFTTRLGRFG-------------------