For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRYAAARIGQSLLVLLLAFTVIFWGVSILPTDPVSIFVAKGDGYFNPEIVEQVKEFYGYDRPAYVQYFSQ LNQLLHGQFGFSLSSGQAVTDRIGSVIGETLKLAGTATVLALAFALATLLLASTTRFEWLRTFIRSIPPL FSAIPTFWLGLVVLQIFSVQLGLFSLFPDGSLASLLVPATVLAVPISAPIAQVLLKNAEATLALPHINTA RAKGGSPTWVIRRHVLKNAAGPALTVTATTIGALLGGSVVTETVFSRSGLGTVLLQAVSNQDISLIQGLV LLTSVVIVGVNLLVDLLYPVLDPRVTKSQRQGFSTRLGRFT*
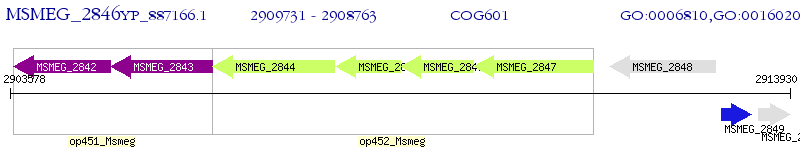
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2846 | - | - | 100% (322) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_4100 | - | 1e-60 | 39.12% (317) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_6866 | - | 4e-47 | 37.58% (306) | dipeptide transport system permease protein DppB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3689c | dppB | 7e-35 | 31.48% (305) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2760 | - | 1e-43 | 35.03% (314) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3665c | dppB | 7e-35 | 31.48% (305) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01124 | - | 1e-18 | 23.33% (330) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0427 | - | 7e-36 | 31.17% (308) | peptide ABC transporter transmembrane protein |
| M. marinum M | MMAR_5153 | dppB | 1e-34 | 32.14% (308) | dipeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_3420 | - | 1e-138 | 76.32% (321) | ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_2645 | - | 1e-39 | 32.37% (312) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4239 | dppB | 1e-34 | 32.14% (308) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3774 | - | 2e-43 | 34.71% (314) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3689c|M.bovis_AF2122/97 ----------------MGWYVARRVAVMVPVFLGATLLIYGMVFLLPGDP
Rv3665c|M.tuberculosis_H37Rv ----------------MGWYVARRVAVMVPVFLGATLLIYGMVFLLPGDP
MMAR_5153|M.marinum_M ----------------MVWYVARRILAMVPVFLGATLLIYGMVFLLPGDP
MUL_4239|M.ulcerans_Agy99 ----------------MVWYVARRILAMVPVFLGATLLIYGMVFLLPGDP
MAB_0427|M.abscessus_ATCC_1997 MTTVLGNVRQFWQSAGLFRYIAKRLLLAIPVLIGTSLLIYSLVYALPGDP
MLBr_01124|M.leprae_Br4923 ----------------MMRFLARRLLNYVVLLALASFLTY-CLTSMAFAP
Mflv_2760|M.gilvum_PYR-GCK --------------MPILRFVARRLLYSAVVMFGVLLVVFALVHLVPGDP
Mvan_3774|M.vanbaalenii_PYR-1 ----------MSTAMPVLRFVARRLLFSAVVMIGVLVVVFALVHLVPGDP
TH_2645|M.thermoresistible__bu ---------MRFVTHPIARFLGRRLLYSLAVLFGVLVVVFGLMQLVPGDP
MSMEG_2846|M.smegmatis_MC2_155 ----------------MLRYAAARIGQSLLVLLLAFTVIFWGVSILPTDP
MAV_3420|M.avium_104 ----------------MLGYVLARIGQSAIVLLAVFSLVFWGVSILPADP
: : *: :: . : : : :. *
Mb3689c|M.bovis_AF2122/97 VAALAGDRP--LTPAVAAQLRSHYH-LDDPFLVQYLRYLGGILHGDLGRA
Rv3665c|M.tuberculosis_H37Rv VAALAGDRP--LTPAVAAQLRSHYH-LDDPFLVQYLRYLGGILHGDLGRA
MMAR_5153|M.marinum_M LAAIAGDRP--LTPAVAQQLRARYH-LDDPFLVQYAHYLGGVLHGDLGRA
MUL_4239|M.ulcerans_Agy99 LAAIAGDRP--LTPAVAQQLRARYH-LDDPFLVQYAHYLGGVLHGDLGRA
MAB_0427|M.abscessus_ATCC_1997 IRALAGDRP--FTPEVMAQLRERFH-LNDPFLVRYGKYIVGFLHGDFGTD
MLBr_01124|M.leprae_Br4923 LDSLLQRSP--HPPQAMVDAKAHALGLDKPIPIRYANWASHAIRGDFGTT
Mflv_2760|M.gilvum_PYR-GCK VRIALGTR---YTPEAYEALRAASG-LDRPLVSQFFGYIGSALTGDLGVS
Mvan_3774|M.vanbaalenii_PYR-1 VRIALGTR---YTPEAYEALRAASG-LDRPIVSQFFGYLGSALTGDLGVS
TH_2645|M.thermoresistible__bu VRIALGTR---YTPEAYQALRSASG-LDRPILEQFVGYLTSAVRGDLGVS
MSMEG_2846|M.smegmatis_MC2_155 VSIFVAKGDGYFNPEIVEQVKEFYG-YDRPAYVQYFSQLNQLLHGQFGFS
MAV_3420|M.avium_104 AAIFVAKGEGYFNPDIVAQVKAFYG-YDRPLWVQYFAQLNQVLHGHFGFS
* : : * :: : *.:*
Mb3689c|M.bovis_AF2122/97 YS-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGLRQGGIFDS
Rv3665c|M.tuberculosis_H37Rv YS-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGLRQGGIFDS
MMAR_5153|M.marinum_M YS-GLPVSAVLAHAFPVTIRLALIALAVEAVLGIGFGVIAGLRQGGIFDA
MUL_4239|M.ulcerans_Agy99 YS-GLPVSAVLAHAFPVTIRLAWIALAVEAVLGIGFGVIAGLRQGGIFDA
MAB_0427|M.abscessus_ATCC_1997 FQ-QRPVAQTVSQRLPVTLRLTVVAVAFETIIGITAGVLCAVRRGSFYDN
MLBr_01124|M.leprae_Br4923 IT-GQPVGSTLGRRVGVSLRLLIIGSLTGTVLGVTAGAWGAIRQYQLSDR
Mflv_2760|M.gilvum_PYR-GCK FRNGDPVTVTLLERLPATLSLGLAGIVIALAIALPAGIYAALREGRLSDA
Mvan_3774|M.vanbaalenii_PYR-1 FRNGDPVTLTLLERLPATLSLGLVGIVIALAIALPAGIYAALREGRLSDA
TH_2645|M.thermoresistible__bu FRNGDPVTLVLLERLPATVSLALAGIAIALAIALPAGIWSALRDGRAGAA
MSMEG_2846|M.smegmatis_MC2_155 LSSGQAVTDRIGSVIGETLKLAGTATVLALAFALATLLLASTTRFEWLRT
MAV_3420|M.avium_104 LSSGQAVTDRIGGVIGETLKLAATATAFAVLFAVSVTALATT--CAPVRS
.* : . :: * . :.:
Mb3689c|M.bovis_AF2122/97 AVLVTGLVIIAIPIFVLG-------FLAQFLFGVQLE----------IAP
Rv3665c|M.tuberculosis_H37Rv AVLVTGLVIIAIPIFVLG-------FLAQFLFGVQLE----------IAP
MMAR_5153|M.marinum_M TVLLTGLIIIAVPIFVLG-------FLAQYLFGVRLG----------LAP
MUL_4239|M.ulcerans_Agy99 TVLLTGLIIIAAPIFVLG-------FLAQYLFGVRLG----------LAP
MAB_0427|M.abscessus_ATCC_1997 LVLVTTTLLVSMPVFVLG-------FLAQYLFGFKLG----------WFP
MLBr_01124|M.leprae_Br4923 VVTLLALLVLSTPTFVIANLLILSALRVNWAFGIQLFDYTGETSPGVYGG
Mflv_2760|M.gilvum_PYR-GCK LVRISSQFGVSIPDFWMG-------ILLIALFSTVLG----WLPTSGYRP
Mvan_3774|M.vanbaalenii_PYR-1 LVRISSQFGVSIPDFWMG-------ILLIALFSTVLG----WLPTSGYRP
TH_2645|M.thermoresistible__bu AVRVASQVGVSVPDFWMG-------ILLIALFASTLG----WLPTSGYRA
MSMEG_2846|M.smegmatis_MC2_155 FIRSIPPLFSAIPTFWLG-------LVVLQIFSVQLG------------L
MAV_3420|M.avium_104 VLRAIPPLFGAVPTFWLG-------LVVLQVFSVQLG------------L
: . : * * :. : *. *
Mb3689c|M.bovis_AF2122/97 VTVGERASVGRLLLPGIVLGAMSFAYVVRLTRSAVAANAHADYVRTATAK
Rv3665c|M.tuberculosis_H37Rv VTVGERASVGRLLLPGIVLGAMSFAYVVRLTRSAVAANAHADYVRTATAK
MMAR_5153|M.marinum_M VTVGARTTLDRLLLPGMVLGAVSFAYVVRLTRSAVAANAHADYVRTATAK
MUL_4239|M.ulcerans_Agy99 VTVGARTTLDRLLLPGMVLGAVSFAYVVRLTRSAVAANAHADYVRTATAK
MAB_0427|M.abscessus_ATCC_1997 I-AGIRDGWYSFLLPGLVLASLSMAYVARLTRTSMLETMDAEFVRTARAK
MLBr_01124|M.leprae_Br4923 AWAHVIDRLRHLILPTLTLALVGAAGYSRYQRNAMLDVLSQDFIRAARAK
Mflv_2760|M.gilvum_PYR-GCK LFEDPAGWLRHIILPALTVAVVAAAIMTRYVRAAVLEVASMGYVRTARSK
Mvan_3774|M.vanbaalenii_PYR-1 LTEDPAGWLRHVVLPGLTVGVVAAAIMTRYVRSAVLEVAAMGYVRTARSK
TH_2645|M.thermoresistible__bu LSDDPVGWLRHLVLPALTVGLVAAAIMTRYIRSAVLEVAAAGHVRTARSK
MSMEG_2846|M.smegmatis_MC2_155 FSLFPDGSLASLLVPATVLAVPISAPIAQVLLKNAEATLALPHINTARAK
MAV_3420|M.avium_104 ISLFPDGSVTSLLVAALVLAVPVSAPIAQVLGKNIDATLALPHVNTARAK
.::. .:. * : .:.:* :*
Mb3689c|M.bovis_AF2122/97 GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAIVTEGIFNIHGVGGV
Rv3665c|M.tuberculosis_H37Rv GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAIVTEGIFNIHGVGGV
MMAR_5153|M.marinum_M GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAVVTEGIFNIHGVGGV
MUL_4239|M.ulcerans_Agy99 GLSRPRVVTVHILRNSLIPVVTFLGADLGALMGGAVVTEGIFNIHGVGGV
MAB_0427|M.abscessus_ATCC_1997 GISESRILLRHTLRNSLIPVVTFIGADVGALLAGAVVTESVFNIPGLGRA
MLBr_01124|M.leprae_Br4923 GLTRRRALLKHGLRTALIPMATLFAYGVAGLVTGALFVEKIFGWHGMGEW
Mflv_2760|M.gilvum_PYR-GCK GLTPRVVTFRHTVRNALVPILTITGIQLATLLGGVIVVEVVFAWPGLGRL
Mvan_3774|M.vanbaalenii_PYR-1 GLSPRIVTFRHTVRNALVPILTITGIQLATLLGGVIVVEVVFAWPGLGRL
TH_2645|M.thermoresistible__bu GLSPWVLTVRHTVRNALIPVLTITGIQLATILGGVIVVEVVFAWPGLGRL
MSMEG_2846|M.smegmatis_MC2_155 GGSPTWVIRRHVLKNAAGPALTVTATTIGALLGGSVVTETVFSRSGLGTV
MAV_3420|M.avium_104 GGTPGWVIRKHVVKNAAGPALTVTATTVGALLGGSVVTETVFSRSGVGAV
* : * ::.: * *. . :. :: * :..* :* *:*
Mb3689c|M.bovis_AF2122/97 LYQAVTRQETPTVVSIVTVLVLIYLITNLLVDLLYAALDPRIRYG-----
Rv3665c|M.tuberculosis_H37Rv LYQAVTRQETPTVVSIVTVLVLIYLITNLLVDLLYAALDPRIRYG-----
MMAR_5153|M.marinum_M LYQAVTRQEAPTVVSIVTVLVMVYLLTNLLVDLLYAALDPRIRYG-----
MUL_4239|M.ulcerans_Agy99 LYQAVTRQEAPTVVSIVTVLVMVYLLTNLLVDLLYAALDPRIRYG-----
MAB_0427|M.abscessus_ATCC_1997 TFDAVRNQEGAVVVSILTLIVFFYIFFNLVVDILYALLDPRIRYE-----
MLBr_01124|M.leprae_Br4923 MVQGVATQDTNIIAAITIFFGTVILLAGLLSDVIYAALDPRVRVS-----
Mflv_2760|M.gilvum_PYR-GCK VFNAVAARDYPLIQGAVLLIAALFLLINLLVDVLYAVVDPRIRLG-----
Mvan_3774|M.vanbaalenii_PYR-1 VFNAVAARDYPLIQGAVLLIAALFLVINLLVDVLYAVVDPRIRLG-----
TH_2645|M.thermoresistible__bu VYNAVAARDYPVIQGAVLLIAALFLLINLLVDLLYAIVDPRIRLA-----
MSMEG_2846|M.smegmatis_MC2_155 LLQAVSNQDISLIQGLVLLTSVVIVGVNLLVDLLYPVLDPRVTKSQRQGF
MAV_3420|M.avium_104 LLQAVSSQDISLIQGLVLLTAVAIVAANLAVDLIYPLLDPRVTRVQRRGF
:.* :: : . . : .* *::*. :***:
Mb3689c|M.bovis_AF2122/97 --------
Rv3665c|M.tuberculosis_H37Rv --------
MMAR_5153|M.marinum_M --------
MUL_4239|M.ulcerans_Agy99 --------
MAB_0427|M.abscessus_ATCC_1997 --------
MLBr_01124|M.leprae_Br4923 --------
Mflv_2760|M.gilvum_PYR-GCK --------
Mvan_3774|M.vanbaalenii_PYR-1 --------
TH_2645|M.thermoresistible__bu --------
MSMEG_2846|M.smegmatis_MC2_155 STRLGRFT
MAV_3420|M.avium_104 TTRLGRFG