For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLIVDTSAPVATPEAPEPGAATSPVRKTLRWVGSHRSLVVSLTIFGVALAWALVPGLFTQYDPISIDNSQ KLLPPSAEHWFGTDLLGRDLYARIVYGARASLLGASMAVAVAVVVGSLLGAVAGWFGGFGDTVAMRFVDV LLSIPGFLLAIAIVVLYTARTGSAGLVPAAVAVGLTSSATFARLIRSEVLRVRASNYVEAAITSGATTVT ILRRHVIPNSLAPTVSLIAVQLGIAIIWIASLSFLGLGAQPPEPEWGLLVSDGRKYIATRGWLTLFPALT IIAVVLATNNISRHLSNRATR*
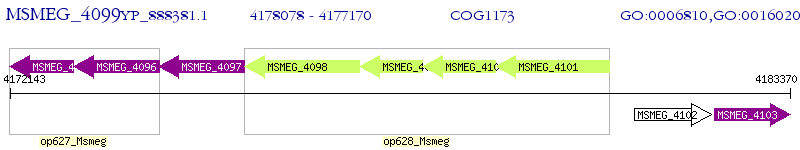
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4099 | - | - | 100% (302) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_2845 | - | 4e-64 | 45.70% (256) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_4387 | - | 1e-59 | 40.42% (287) | ABC transporter permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3688c | dppC | 2e-41 | 40.32% (253) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2759 | - | 2e-47 | 38.34% (253) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3664c | dppC | 2e-41 | 40.32% (253) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01123 | - | 9e-31 | 29.21% (267) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 3e-43 | 36.90% (290) | peptide ABC transporter DppC |
| M. marinum M | MMAR_5152 | dppC | 3e-42 | 38.80% (250) | dipeptide-transport integral membrane protein ABC |
| M. avium 104 | MAV_3419 | - | 2e-61 | 45.28% (254) | ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_2705 | - | 2e-49 | 38.46% (273) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4238 | dppC | 3e-42 | 38.80% (250) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3775 | - | 1e-47 | 39.29% (252) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2759|M.gilvum_PYR-GCK --------MTTTD---PQS------TRLSSWRLLALNPVTLISALVLIAV
Mvan_3775|M.vanbaalenii_PYR-1 --------MTTTD---PND------TRLSSWRLLAGNPVTLVSALVLVAV
TH_2705|M.thermoresistible__bu --------MTADD---LTGGEPDAVSRVAAWRLLAANPVTLLSAALLALL
MLBr_01123|M.leprae_Br4923 --------MTLTEGLSISEKTNFASRKTLVLRRFAHNRTASVSLMLLVLL
MSMEG_4099|M.smegmatis_MC2_155 MSLIVDTSAPVATPEAPEP--GAATSPVRKTLRWVGSHRSLVVSLTIFGV
MAV_3419|M.avium_104 ------MSARTAS------------WPVR-------------ISAAVLIL
Mb3688c|M.bovis_AF2122/97 -----------------------------------------MIAAALILL
Rv3664c|M.tuberculosis_H37Rv -----------------------------------------MIAAALILL
MMAR_5152|M.marinum_M --------------------MAELGGFWFDILGGLRRRPKFVIAAVLIVF
MUL_4238|M.ulcerans_Agy99 --------------------MAELRGFWFDILGGLRRRPKFVIAAVLIVF
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRSGFFITGSILLTI
: .
Mflv_2759|M.gilvum_PYR-GCK AVVAVTANWIIP--YGVNDVDVPNALAPPSGSHWFGTDELGRDVLSRVLV
Mvan_3775|M.vanbaalenii_PYR-1 TVVAVTANWLAP--YGVNDVDVPNALAPPSASHWFGTDELGRDVLSRVLV
TH_2705|M.thermoresistible__bu VAVAVTAPWLAP--YGVNDINVPEALQPPSAAHWFGTDELGRDVLSRVLV
MLBr_01123|M.leprae_Br4923 FVACYLLPSLLP--YSYDDLDFNALLQPPNTRHWLGTNALGQDLLAQTLR
MSMEG_4099|M.smegmatis_MC2_155 ALAWALVPGLFT-QYDPISIDNSQKLLPPSAEHWFGTDLLGRDLYARIVY
MAV_3419|M.avium_104 TAAWAAAPGLFT-DRNPLKGRPVDKFQPPSAAHWFGTDHLGRDVLTRVIY
Mb3688c|M.bovis_AF2122/97 ILVVAAFPSLFT-AADPTYADPSQSMLAPSAAHWFGTDLQGHDIYSRTVY
Rv3664c|M.tuberculosis_H37Rv ILVVAAFPSLFT-AADPTYADPSQSMLAPSAAHWFGTDLQGHDIYSRTVY
MMAR_5152|M.marinum_M ILVVAAFPSLFT-GIDPSYADPSQSMLSPSTAHWFGTDLQGHDIYARTVY
MUL_4238|M.ulcerans_Agy99 ILFVAAFPSLFT-GIDPSYADPSQSMLSPSTAHWFGTDLQGHDIYARTVY
MAB_0428|M.abscessus_ATCC_1997 LVLMALAPQLFTFGKDPRACDLYQARKGISAAHWLGTDLQGCDYYANVVY
: . . . **:**: * * :. :
Mflv_2759|M.gilvum_PYR-GCK ATQASMRIAVVSVAFAVIVGVAVGVIAGYRGGWLDTVLMRVVDVMFAFP-
Mvan_3775|M.vanbaalenii_PYR-1 AVQASMRIAAVSVALAMVVGVAVGVVAGYRGGWLDTVLMRVVDVMFAFP-
TH_2705|M.thermoresistible__bu AIQASLRVAAVSVAFAAAVGILVGLLAGYRGGWLDTVLMRLVDVLFAFP-
MLBr_01123|M.leprae_Br4923 GMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWVVDLLLVVPS
MSMEG_4099|M.smegmatis_MC2_155 GARASLLGASMAVAVAVVVGSLLGAVAGWFGGFGDTVAMRFVDVLLSIPG
MAV_3419|M.avium_104 GTSHTVATAGLAVAVGLLLGSSVGIAAGVSGPVVDAVAMRASDVLLALPG
Mb3688c|M.bovis_AF2122/97 GARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFLGLPL
Rv3664c|M.tuberculosis_H37Rv GARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFLGLPL
MMAR_5152|M.marinum_M GARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGLPL
MUL_4238|M.ulcerans_Agy99 GARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGLPL
MAB_0428|M.abscessus_ATCC_1997 GARVSLTIGLIAMVGVLIIGVIVGALAGYFGGIADSALSRLTDVFYGIPT
. :: . . :* ** * * . *:: .*
Mflv_2759|M.gilvum_PYR-GCK -VLLLALAVVAILGP---GVTTTILAIGIVYTPIFARVARASTLGVRTEP
Mvan_3775|M.vanbaalenii_PYR-1 -VLLLALAVVAILGP---GVTTTILAIAIVYTPIFARVARASTLGVRTEP
TH_2705|M.thermoresistible__bu -VLLLALAIVAILGP---GMTTTMLAIGIVFTPIFARVARAGTLGVRVEP
MLBr_01123|M.leprae_Br4923 FVLIAIMSPQTKSSD---NIMLLVLLLAGFGWMVSSRMVRGMTMSLRKSE
MSMEG_4099|M.smegmatis_MC2_155 FLLAIAIVVLYTARTGSAGLVPAAVAVGLTSSATFARLIRSEVLRVRASN
MAV_3419|M.avium_104 FLTSVWIVTAYGP-----GPLSVGIGVGIGSIAVFARVFRAEVLRVRALD
Mb3688c|M.bovis_AF2122/97 LLAAIVLMQVMHHR----TVWTVIAILALFGWPQVARIARGAVLEVRASD
Rv3664c|M.tuberculosis_H37Rv LLAAIVLMQVMHHR----TVWTVIAILALFGWPQVARIARGAVLEVRASD
MMAR_5152|M.marinum_M LLAAIVLMQVMHHR----TVWTVIAILALFGWPQIARIARSSVLEVRGSD
MUL_4238|M.ulcerans_Agy99 LLAAIVLMQVMHHR----TVWTVIAILALFGWPQIARIARSSVLEVRGSD
MAB_0428|M.abscessus_ATCC_1997 ILGGMILLSVVGHR----TVWSVAGALIAFGWMTSMRLVRSSVMSIKQSD
: : : *: *. .: ::
Mflv_2759|M.gilvum_PYR-GCK YVSVSRAMGAGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFLG
Mvan_3775|M.vanbaalenii_PYR-1 YVSASRAMGTGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFLG
TH_2705|M.thermoresistible__bu FVQASHTMGTGHAYILRRHILPNIAAPLIVQTSLSLAFAILAEAALSFLG
MLBr_01123|M.leprae_Br4923 FIRAGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSAILAETGLSFLG
MSMEG_4099|M.smegmatis_MC2_155 YVEAAITSGATTVTILRRHVIPNSLAPTVSLIAVQLGIAIIWIASLSFLG
MAV_3419|M.avium_104 YVEAAFLSGETRWSVIRRHIVPNAAGAVIALAVIDLSGAILLISALGYLG
Mb3688c|M.bovis_AF2122/97 YVLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYLG
Rv3664c|M.tuberculosis_H37Rv YVLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYLG
MMAR_5152|M.marinum_M FVLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYLG
MUL_4238|M.ulcerans_Agy99 FVLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYLG
MAB_0428|M.abscessus_ATCC_1997 YVQAAIALGAPTWRILLRHILPNALAPVLVYATIAVGSFIAAEATLTYMG
:: .. * :: * :** . : : :. * : * ::*
Mflv_2759|M.gilvum_PYR-GCK LGIQPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFLIVLAFNLFGDGLR
Mvan_3775|M.vanbaalenii_PYR-1 LGIQPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFVIVLAFNLLGDGLR
TH_2705|M.thermoresistible__bu LGIQPPEPSLGRMIFDSQGFVTLAWWMAVFPGAAIVAAVLACNLFGDGLR
MLBr_01123|M.leprae_Br4923 FGIKPPDVSLGTLIANGTQSATTFPWVFLFPAGVLVVILVCANLTGDGLR
MSMEG_4099|M.smegmatis_MC2_155 LGAQPPEPEWGLLVSDGRKYIATRGWLTLFPALTIIAVVLATNNISRHLS
MAV_3419|M.avium_104 YSAPPPTPEWGLLVAEGRRYLATAWWLSTLPGAVVLSVILALGVLSRRAL
Mb3688c|M.bovis_AF2122/97 VGLPTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDALR
Rv3664c|M.tuberculosis_H37Rv VGLPTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDALR
MMAR_5152|M.marinum_M VGLPPTVVSWGGDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALR
MUL_4238|M.ulcerans_Agy99 VGLPPTVVSWGDDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALR
MAB_0428|M.abscessus_ATCC_1997 IGLELPEISWGLQINDGQSRFASTPRLVIVPALFLSAAVLGFIMVGDALR
. . . * : . : *. : :: .
Mflv_2759|M.gilvum_PYR-GCK DVLDPKQRTTIEARRLQR-------------
Mvan_3775|M.vanbaalenii_PYR-1 DVLDPKQRTTVEARRRER-------------
TH_2705|M.thermoresistible__bu DVLDPKQRTVIETQRRARQRPGRGRRAGSAW
MLBr_01123|M.leprae_Br4923 DAFDPVSKHLRR-------------------
MSMEG_4099|M.smegmatis_MC2_155 NRATR--------------------------
MAV_3419|M.avium_104 TSHRI--------------------------
Mb3688c|M.bovis_AF2122/97 DALDPASRAWRA-------------------
Rv3664c|M.tuberculosis_H37Rv DALDPASRAWRA-------------------
MMAR_5152|M.marinum_M DALDPASRAWRA-------------------
MUL_4238|M.ulcerans_Agy99 DALDPASRAWRA-------------------
MAB_0428|M.abscessus_ATCC_1997 DALDPRRR-----------------------