For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AELGGFWFDILGGLRRRPKFVIAAVLIVFILVVAAFPSLFTGIDPSYADPSQSMLSPSTAHWFGTDLQGH DIYARTVYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGLPLLLAAIVLMQVMH HRTVWTVIAILALFGWPQIARIARSSVLEVRGSDFVLAAKALGMSRFQILLRHALPNAIGPVIAVATVAL GIFIVTEATLSYLGVGLPPTVVSWGGDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALRDALDPA SRAWRA*
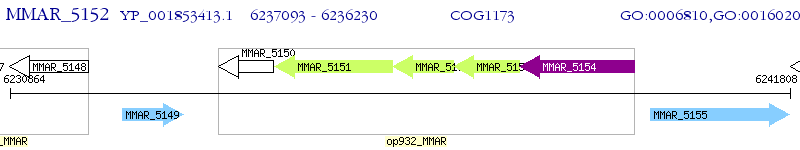
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5152 | dppC | - | 100% (287) | dipeptide-transport integral membrane protein ABC transporter DppC |
| M. marinum M | MMAR_4137 | oppC | 1e-37 | 32.61% (276) | oligopeptide-transport integral membrane protein ABC |
| M. marinum M | MMAR_5153 | dppB | 2e-07 | 23.85% (239) | dipeptide-transport integral membrane protein ABC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3688c | dppC | 1e-132 | 86.84% (266) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2759 | - | 5e-46 | 36.06% (269) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3664c | dppC | 1e-132 | 86.84% (266) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01123 | - | 2e-36 | 32.21% (267) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 4e-72 | 47.54% (284) | peptide ABC transporter DppC |
| M. avium 104 | MAV_0466 | dppC | 1e-135 | 81.98% (283) | ABC transporter, permease protein DppC |
| M. smegmatis MC2 155 | MSMEG_4356 | - | 4e-46 | 36.80% (269) | inner membrane ABC transporter permease protein YddQ |
| M. thermoresistible (build 8) | TH_2705 | - | 2e-42 | 35.32% (269) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4238 | dppC | 1e-159 | 98.95% (287) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3775 | - | 2e-47 | 36.80% (269) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2759|M.gilvum_PYR-GCK ------------MTTTDP------QSTRLSSWRLLALNPVTLISALVLIA
Mvan_3775|M.vanbaalenii_PYR-1 ------------MTTTDP------NDTRLSSWRLLAGNPVTLVSALVLVA
MSMEG_4356|M.smegmatis_MC2_155 ------------MTVT--------EQTRVASWRLLLSNPVTAVSAAVLVV
TH_2705|M.thermoresistible__bu ------------MTADDLTGGEPDAVSRVAAWRLLAANPVTLLSAALLAL
MMAR_5152|M.marinum_M --------------------MAELGGFWFDILGGLRRRPKFVIAAVLIVF
MUL_4238|M.ulcerans_Agy99 --------------------MAELRGFWFDILGGLRRRPKFVIAAVLIVF
Mb3688c|M.bovis_AF2122/97 -----------------------------------------MIAAALILL
Rv3664c|M.tuberculosis_H37Rv -----------------------------------------MIAAALILL
MAV_0466|M.avium_104 ----------------MAERIRARGGFWRETWRRLRRRPKFIGAGLLILV
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRSGFFITGSILLTI
MLBr_01123|M.leprae_Br4923 ------------MTLTEGLSISEKTNFASRKTLVLRRFAHNRTASVSLML
.. :
Mflv_2759|M.gilvum_PYR-GCK VAVVAVTANWII---PYGVNDVDVPNALAPPSGSHWFGTDELGRDVLSRV
Mvan_3775|M.vanbaalenii_PYR-1 VTVVAVTANWLA---PYGVNDVDVPNALAPPSASHWFGTDELGRDVLSRV
MSMEG_4356|M.smegmatis_MC2_155 VAVVALTAQWIA---PFGVNDIDVPSALQGPGGTHWFGTDELGRDVFSRI
TH_2705|M.thermoresistible__bu LVAVAVTAPWLA---PYGVNDINVPEALQPPSAAHWFGTDELGRDVLSRV
MMAR_5152|M.marinum_M ILVVAAFPSLFT---GIDPSYADPSQSMLSPSTAHWFGTDLQGHDIYART
MUL_4238|M.ulcerans_Agy99 ILFVAAFPSLFT---GIDPSYADPSQSMLSPSTAHWFGTDLQGHDIYART
Mb3688c|M.bovis_AF2122/97 ILVVAAFPSLFT---AADPTYADPSQSMLAPSAAHWFGTDLQGHDIYSRT
Rv3664c|M.tuberculosis_H37Rv ILVVAAFPSLFT---AADPTYADPSQSMLAPSAAHWFGTDLQGHDIYSRT
MAV_0466|M.avium_104 ILAVALFPALFT---AADPTYADPAQSLLPPSRTHWFGTDLQGHDVYART
MAB_0428|M.abscessus_ATCC_1997 LVLMALAPQLFTF--GKDPRACDLYQARKGISAAHWLGTDLQGCDYYANV
MLBr_01123|M.leprae_Br4923 LVLLFVACYLLPSLLPYSYDDLDFNALLQPPNTRHWLGTNALGQDLLAQT
: : : . : . **:**: * * :.
Mflv_2759|M.gilvum_PYR-GCK LVATQASMRIAVVSVAFAVIVGVAVGVIAGYRGGWLDTVLMRVVDVMFAF
Mvan_3775|M.vanbaalenii_PYR-1 LVAVQASMRIAAVSVALAMVVGVAVGVVAGYRGGWLDTVLMRVVDVMFAF
MSMEG_4356|M.smegmatis_MC2_155 LVAIQASLRVAVVSVALAAVVGVTIGVVAGYRGGWLDTIVMRVVDVMFAF
TH_2705|M.thermoresistible__bu LVAIQASLRVAAVSVAFAAAVGILVGLLAGYRGGWLDTVLMRLVDVLFAF
MMAR_5152|M.marinum_M VYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGL
MUL_4238|M.ulcerans_Agy99 VYGARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGL
Mb3688c|M.bovis_AF2122/97 VYGARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFLGL
Rv3664c|M.tuberculosis_H37Rv VYGARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFLGL
MAV_0466|M.avium_104 VYGARASVTVGLGAAAIVFVVGGALGALAGFYGGWIDAVVSRVTDVFFGL
MAB_0428|M.abscessus_ATCC_1997 VYGARVSLTIGLIAMVGVLIIGVIVGALAGYFGGIADSALSRLTDVFYGI
MLBr_01123|M.leprae_Br4923 LRGMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWVVDLLLVV
: . : *: :. . :* :**: * * : :.*:: .
Mflv_2759|M.gilvum_PYR-GCK P-VLLLALAVVAILGPGVTTTILAIGIVY-TPIFARVARASTLGVRTEPY
Mvan_3775|M.vanbaalenii_PYR-1 P-VLLLALAVVAILGPGVTTTILAIAIVY-TPIFARVARASTLGVRTEPY
MSMEG_4356|M.smegmatis_MC2_155 P-VLLLALAIVAILGPGVTTTMLAIGIVY-IPIFARVARASTLGVRVEPY
TH_2705|M.thermoresistible__bu P-VLLLALAIVAILGPGMTTTMLAIGIVF-TPIFARVARAGTLGVRVEPF
MMAR_5152|M.marinum_M P-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQIARIARSSVLEVRGSDF
MUL_4238|M.ulcerans_Agy99 P-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQIARIARSSVLEVRGSDF
Mb3688c|M.bovis_AF2122/97 P-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQVARIARGAVLEVRASDY
Rv3664c|M.tuberculosis_H37Rv P-LLLAAIVLMQVMHHRTVWTVIAILALFGWPQVARIARGAVLEVRASDY
MAV_0466|M.avium_104 P-LLLAAIVLMQVLHHRTVWTVIAILALFGWPQVARIARGAVLAVRASDY
MAB_0428|M.abscessus_ATCC_1997 P-TILGGMILLSVVGHRTVWSVAGALIAFGWMTSMRLVRSSVMSIKQSDY
MLBr_01123|M.leprae_Br4923 PSFVLIAIMSPQTKSSDNIMLLVLLLAGFGWMVSSRMVRGMTMSLRKSEF
* :* .: : : *:.*. .: :: . :
Mflv_2759|M.gilvum_PYR-GCK VSVSRAMGAGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFLGL
Mvan_3775|M.vanbaalenii_PYR-1 VSASRAMGTGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFLGL
MSMEG_4356|M.smegmatis_MC2_155 VAVSRTMGTPSGFILRRHILPNIAGPLIVQLSLSLAFAILAEASLSFLGL
TH_2705|M.thermoresistible__bu VQASHTMGTGHAYILRRHILPNIAAPLIVQTSLSLAFAILAEAALSFLGL
MMAR_5152|M.marinum_M VLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYLGV
MUL_4238|M.ulcerans_Agy99 VLAAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYLGV
Mb3688c|M.bovis_AF2122/97 VLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYLGV
Rv3664c|M.tuberculosis_H37Rv VLAAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYLGV
MAV_0466|M.avium_104 VLAAKALGMSRFQILIRHALPNALGPVIAVATIALGLFIVTEATLSYLGV
MAB_0428|M.abscessus_ATCC_1997 VQAAIALGAPTWRILLRHILPNALAPVLVYATIAVGSFIAAEATLTYMGI
MLBr_01123|M.leprae_Br4923 IRAGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSAILAETGLSFLGF
: .. :* :: * :** . :: :: :. * :*: *:::*.
Mflv_2759|M.gilvum_PYR-GCK GIQPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFLIVLAFNLFGDGLRD
Mvan_3775|M.vanbaalenii_PYR-1 GIQPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFVIVLAFNLLGDGLRD
MSMEG_4356|M.smegmatis_MC2_155 GIQPPQPSLGRMIFDAQGFVTLAWWMAVFPGAAIFVTVLAFNLFGDGLRD
TH_2705|M.thermoresistible__bu GIQPPEPSLGRMIFDSQGFVTLAWWMAVFPGAAIVAAVLACNLFGDGLRD
MMAR_5152|M.marinum_M GLPPTVVSWGGDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALRD
MUL_4238|M.ulcerans_Agy99 GLPPTVVSWGDDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALRD
Mb3688c|M.bovis_AF2122/97 GLPTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDALRD
Rv3664c|M.tuberculosis_H37Rv GLPTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDALRD
MAV_0466|M.avium_104 GLPPSVVSWGGDINLAQTRLRAGSPILFYPAGALAATVLAFMMMGDALRD
MAB_0428|M.abscessus_ATCC_1997 GLELPEISWGLQINDGQSRFASTPRLVIVPALFLSAAVLGFIMVGDALRD
MLBr_01123|M.leprae_Br4923 GIKPPDVSLGTLIANGTQSATTFPWVFLFPAGVLVVILVCANLTGDGLRD
*: . * * * . : . *. : :: : **.***
Mflv_2759|M.gilvum_PYR-GCK VLDPKQRTTIEARRLQR-------------
Mvan_3775|M.vanbaalenii_PYR-1 VLDPKQRTTVEARRRER-------------
MSMEG_4356|M.smegmatis_MC2_155 VLDPKQRTLMEAKRTRQ-------------
TH_2705|M.thermoresistible__bu VLDPKQRTVIETQRRARQRPGRGRRAGSAW
MMAR_5152|M.marinum_M ALDPASRAWRA-------------------
MUL_4238|M.ulcerans_Agy99 ALDPASRAWRA-------------------
Mb3688c|M.bovis_AF2122/97 ALDPASRAWRA-------------------
Rv3664c|M.tuberculosis_H37Rv ALDPASRAWRA-------------------
MAV_0466|M.avium_104 ALDPASRAWRA-------------------
MAB_0428|M.abscessus_ATCC_1997 ALDPRRR-----------------------
MLBr_01123|M.leprae_Br4923 AFDPVSKHLRR-------------------
.:** :