For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SARTASWPVRISAAVLILTAAWAAAPGLFTDRNPLKGRPVDKFQPPSAAHWFGTDHLGRDVLTRVIYGTS HTVATAGLAVAVGLLLGSSVGIAAGVSGPVVDAVAMRASDVLLALPGFLTSVWIVTAYGPGPLSVGIGVG IGSIAVFARVFRAEVLRVRALDYVEAAFLSGETRWSVIRRHIVPNAAGAVIALAVIDLSGAILLISALGY LGYSAPPPTPEWGLLVAEGRRYLATAWWLSTLPGAVVLSVILALGVLSRRALTSHRI*
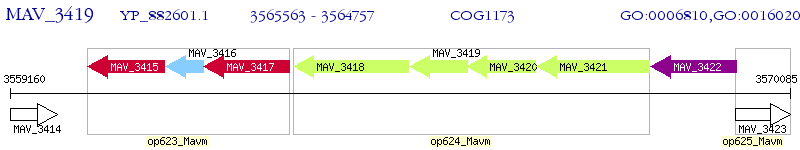
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3419 | - | - | 100% (268) | ABC transporter permease protein |
| M. avium 104 | MAV_0466 | dppC | 1e-32 | 38.29% (222) | ABC transporter, permease protein DppC |
| M. avium 104 | MAV_1432 | oppC | 2e-23 | 29.58% (213) | ABC transporter, permease protein OppC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3688c | dppC | 8e-33 | 37.90% (219) | peptide ABC transporter transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2759 | - | 6e-44 | 37.10% (248) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3664c | dppC | 8e-33 | 37.90% (219) | peptide ABC transporter transmembrane protein |
| M. leprae Br4923 | MLBr_01123 | - | 1e-21 | 27.78% (216) | putative binding-protein dependent transport protein |
| M. abscessus ATCC 19977 | MAB_0428 | - | 2e-25 | 31.30% (262) | peptide ABC transporter DppC |
| M. marinum M | MMAR_5152 | dppC | 9e-31 | 34.86% (218) | dipeptide-transport integral membrane protein ABC |
| M. smegmatis MC2 155 | MSMEG_2845 | - | 1e-122 | 81.13% (265) | ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_2705 | - | 1e-44 | 37.36% (265) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_4238 | dppC | 5e-31 | 36.04% (222) | dipeptide-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3775 | - | 1e-44 | 36.95% (249) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3688c|M.bovis_AF2122/97 -----------------------------------------MIAAALILL
Rv3664c|M.tuberculosis_H37Rv -----------------------------------------MIAAALILL
MMAR_5152|M.marinum_M --------------------MAELGGFWFDILGGLRRRPKFVIAAVLIVF
MUL_4238|M.ulcerans_Agy99 --------------------MAELRGFWFDILGGLRRRPKFVIAAVLIVF
MAB_0428|M.abscessus_ATCC_1997 MSDPVNTPGPVSGPDATAESLAEQADLWGNAWKYLRRSGFFITGSILLTI
Mflv_2759|M.gilvum_PYR-GCK ------------MTTTDPQS------TRLSSWRLLALNPVTLISALVLIA
Mvan_3775|M.vanbaalenii_PYR-1 ------------MTTTDPND------TRLSSWRLLAGNPVTLVSALVLVA
TH_2705|M.thermoresistible__bu ------------MTADDLTGGEPDAVSRVAAWRLLAANPVTLLSAALLAL
MAV_3419|M.avium_104 -------------------------------MSARTASWPVRISAAVLIL
MSMEG_2845|M.smegmatis_MC2_155 ---------MSITVGPPSDHPDTETRRNKSLSALRAAPWSARISGALLVL
MLBr_01123|M.leprae_Br4923 ---------MTLTEGLSISEKTNFASRKTLVLRRFAHNRTASVSLMLLVL
. ::
Mb3688c|M.bovis_AF2122/97 ILVVAAFPSLFT-AADPTYADPSQSMLAPSAAHWFGTDLQGHDIYSRTVY
Rv3664c|M.tuberculosis_H37Rv ILVVAAFPSLFT-AADPTYADPSQSMLAPSAAHWFGTDLQGHDIYSRTVY
MMAR_5152|M.marinum_M ILVVAAFPSLFT-GIDPSYADPSQSMLSPSTAHWFGTDLQGHDIYARTVY
MUL_4238|M.ulcerans_Agy99 ILFVAAFPSLFT-GIDPSYADPSQSMLSPSTAHWFGTDLQGHDIYARTVY
MAB_0428|M.abscessus_ATCC_1997 LVLMALAPQLFTFGKDPRACDLYQARKGISAAHWLGTDLQGCDYYANVVY
Mflv_2759|M.gilvum_PYR-GCK VAVVAVTANWII-PYGVNDVDVPNALAPPSGSHWFGTDELGRDVLSRVLV
Mvan_3775|M.vanbaalenii_PYR-1 VTVVAVTANWLA-PYGVNDVDVPNALAPPSASHWFGTDELGRDVLSRVLV
TH_2705|M.thermoresistible__bu LVAVAVTAPWLA-PYGVNDINVPEALQPPSAAHWFGTDELGRDVLSRVLV
MAV_3419|M.avium_104 TAAWAAAPGLFT-DRNPLKGRPVDKFQPPSAAHWFGTDHLGRDVLTRVIY
MSMEG_2845|M.smegmatis_MC2_155 VAAWAVAPGVFT-SADPLNGNRLEKFQPPSWAHWFGTDHLGRDVLTRVIH
MLBr_01123|M.leprae_Br4923 LFVACYLLPSLL-PYSYDDLDFNALLQPPNTRHWLGTNALGQDLLAQTLR
. : . . **:**: * * :..:
Mb3688c|M.bovis_AF2122/97 GARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFLGLP-
Rv3664c|M.tuberculosis_H37Rv GARASVTVGLGATLAVFVVGGALGALAGFYGSWIDAVVSRVTDVFLGLP-
MMAR_5152|M.marinum_M GARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGLP-
MUL_4238|M.ulcerans_Agy99 GARASVVVGLGATAAAFIAGVTLGALAGYYGGWLDAVVSRITDIFFGLP-
MAB_0428|M.abscessus_ATCC_1997 GARVSLTIGLIAMVGVLIIGVIVGALAGYFGGIADSALSRLTDVFYGIP-
Mflv_2759|M.gilvum_PYR-GCK ATQASMRIAVVSVAFAVIVGVAVGVIAGYRGGWLDTVLMRVVDVMFAFP-
Mvan_3775|M.vanbaalenii_PYR-1 AVQASMRIAAVSVALAMVVGVAVGVVAGYRGGWLDTVLMRVVDVMFAFP-
TH_2705|M.thermoresistible__bu AIQASLRVAAVSVAFAAAVGILVGLLAGYRGGWLDTVLMRLVDVLFAFP-
MAV_3419|M.avium_104 GTSHTVATAGLAVAVGLLLGSSVGIAAGVSGPVVDAVAMRASDVLLALP-
MSMEG_2845|M.smegmatis_MC2_155 GTSHTLLTAGLAVAVGLVVGSIIGILAGVSGPVADAVGMRLSDVLLALP-
MLBr_01123|M.leprae_Br4923 GMQKSMLIGVCVALISTSIAATVGSIAGYFGRWRDRVLMWVVDLLLVVPS
. :: . . :* ** * * . *:: .*
Mb3688c|M.bovis_AF2122/97 LLLAAIVLMQVMHHRTVWTVIAILALFGWPQVARIARGAVLEVRASDYVL
Rv3664c|M.tuberculosis_H37Rv LLLAAIVLMQVMHHRTVWTVIAILALFGWPQVARIARGAVLEVRASDYVL
MMAR_5152|M.marinum_M LLLAAIVLMQVMHHRTVWTVIAILALFGWPQIARIARSSVLEVRGSDFVL
MUL_4238|M.ulcerans_Agy99 LLLAAIVLMQVMHHRTVWTVIAILALFGWPQIARIARSSVLEVRGSDFVL
MAB_0428|M.abscessus_ATCC_1997 TILGGMILLSVVGHRTVWSVAGALIAFGWMTSMRLVRSSVMSIKQSDYVQ
Mflv_2759|M.gilvum_PYR-GCK VLLLALAVVAILGPGVTTTILAIGIVY-TPIFARVARASTLGVRTEPYVS
Mvan_3775|M.vanbaalenii_PYR-1 VLLLALAVVAILGPGVTTTILAIAIVY-TPIFARVARASTLGVRTEPYVS
TH_2705|M.thermoresistible__bu VLLLALAIVAILGPGMTTTMLAIGIVF-TPIFARVARAGTLGVRVEPFVQ
MAV_3419|M.avium_104 GFLTSVWIVTAYGPGPLSVGIGVGIGS-IAVFARVFRAEVLRVRALDYVE
MSMEG_2845|M.smegmatis_MC2_155 GFLVSVWIVTAYGAGPLSVGIGVGIGS-IAIFARVFRAEVMRVRALDYVE
MLBr_01123|M.leprae_Br4923 FVLIAIMSPQTKSSDNIMLLVLLLAGFGWMVSSRMVRGMTMSLRKSEFIR
.* .: *: *. .: :: ::
Mb3688c|M.bovis_AF2122/97 AAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYLGVGL
Rv3664c|M.tuberculosis_H37Rv AAKALGLNRFQILLRHALPNAVGPVIAVATVALGIFIVTEATLSYLGVGL
MMAR_5152|M.marinum_M AAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYLGVGL
MUL_4238|M.ulcerans_Agy99 AAKALGMSRFQILLRHALPNAIGPVIAVATVALGIFIVTEATLSYLGVGL
MAB_0428|M.abscessus_ATCC_1997 AAIALGAPTWRILLRHILPNALAPVLVYATIAVGSFIAAEATLTYMGIGL
Mflv_2759|M.gilvum_PYR-GCK VSRAMGAGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFLGLGI
Mvan_3775|M.vanbaalenii_PYR-1 ASRAMGTGDLYILRRHILPNIAGPLIVQTSLSLAFAILSEAALSFLGLGI
TH_2705|M.thermoresistible__bu ASHTMGTGHAYILRRHILPNIAAPLIVQTSLSLAFAILAEAALSFLGLGI
MAV_3419|M.avium_104 AAFLSGETRWSVIRRHIVPNAAGAVIALAVIDLSGAILLISALGYLGYSA
MSMEG_2845|M.smegmatis_MC2_155 AAFLSGESRWSVIRRHIVPNAIGAVVALAVIDLSAAILAISALGYLGYSA
MLBr_01123|M.leprae_Br4923 AGKYMGVSSRRMIVGHVVPNVASILIIDATLNVGSAILAETGLSFLGFGI
.. * :: * :** . :: : : :. * : * ::* .
Mb3688c|M.bovis_AF2122/97 PTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDALRDAL
Rv3664c|M.tuberculosis_H37Rv PTSVVSWGGDINVAQTRLRSGSPILFYPAGALAITVLAFMMMGDALRDAL
MMAR_5152|M.marinum_M PPTVVSWGGDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALRDAL
MUL_4238|M.ulcerans_Agy99 PPTVVSWGDDINVAQMRLRAGSPILFYPAGALAVTVLAFMMMGDALRDAL
MAB_0428|M.abscessus_ATCC_1997 ELPEISWGLQINDGQSRFASTPRLVIVPALFLSAAVLGFIMVGDALRDAL
Mflv_2759|M.gilvum_PYR-GCK QPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFLIVLAFNLFGDGLRDVL
Mvan_3775|M.vanbaalenii_PYR-1 QPPQPSLGRMIFDSQGFVTMAWWMAVFPGAAIFVIVLAFNLLGDGLRDVL
TH_2705|M.thermoresistible__bu QPPEPSLGRMIFDSQGFVTLAWWMAVFPGAAIVAAVLACNLFGDGLRDVL
MAV_3419|M.avium_104 PPPTPEWGLLVAEGRRYLATAWWLSTLPGAVVLSVILALGVLS---RRAL
MSMEG_2845|M.smegmatis_MC2_155 PPPTPEWGLLVAEGRSYLATAWWLTSLPGAVILVVIVALGVVS---RRVL
MLBr_01123|M.leprae_Br4923 KPPDVSLGTLIANGTQSATTFPWVFLFPAGVLVVILVCANLTGDGLRDAF
. . * : . : *. : :: : . * .:
Mb3688c|M.bovis_AF2122/97 DPASRAWRA-------------------
Rv3664c|M.tuberculosis_H37Rv DPASRAWRA-------------------
MMAR_5152|M.marinum_M DPASRAWRA-------------------
MUL_4238|M.ulcerans_Agy99 DPASRAWRA-------------------
MAB_0428|M.abscessus_ATCC_1997 DPRRR-----------------------
Mflv_2759|M.gilvum_PYR-GCK DPKQRTTIEARRLQR-------------
Mvan_3775|M.vanbaalenii_PYR-1 DPKQRTTVEARRRER-------------
TH_2705|M.thermoresistible__bu DPKQRTVIETQRRARQRPGRGRRAGSAW
MAV_3419|M.avium_104 TSHRI-----------------------
MSMEG_2845|M.smegmatis_MC2_155 KSNNI-----------------------
MLBr_01123|M.leprae_Br4923 DPVSKHLRR-------------------
.