For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSASRSLAAPLLKVDAVTKRYIGVTALTDASLAVGAGEVRALLGRNGAGKSTLIRILSGVEQADTGTVSI DGTILDGGGVRRANELGVQTVHQELSLVPDMTVAENMFVGAWPRSGGKIDYDAMRAEASTVIDRLGLDID PDSPVAALSLATRQLVEICRAVRRQPRVLILDEPTSALAATEVSIVLDTVTRIAQTGVAVIYVSHRLDEI RRIASSATILRDGRVVDTVEMDTTGTDAMIAMMVGPDHQAQERPPLRAVDRTRTPLLSVRDLRLPPKIEK VSFDLFAGEVLGIAGLMGSGRTEVLRAVAGFDTPVSGEMTLDGKPMPWNAPAVMKRNGVGLTPEDRKHDA IVPLLGVDENMVLSDYTRVRHHGPISLRRMRTAAQSLIDRLRIETASPVTPIVNLSGGNQQKAIIGRWLH ADASILLLDEPTRGVDVAAKAGIYDLVRALADAGKAVIFVSGELEELPLVCDRVITVQGGRVTAEFIGVD VTVDAVLSAAMAA
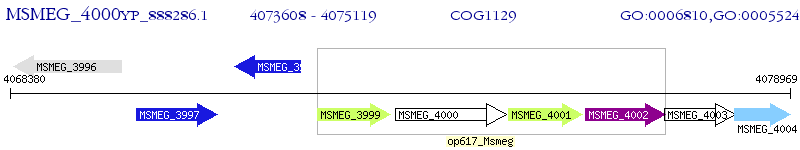
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4000 | - | - | 100% (503) | ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_4170 | - | 8e-91 | 38.58% (508) | ribose transport ATP-binding protein RbsA |
| M. smegmatis MC2 155 | MSMEG_1711 | - | 2e-86 | 37.68% (475) | ATP binding protein of ABC transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3687c | dppD | 3e-28 | 26.40% (519) | peptide ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_2607 | - | 3e-35 | 36.36% (253) | ABC transporter related |
| M. tuberculosis H37Rv | Rv3663c | dppD | 3e-28 | 26.40% (519) | peptide ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_00848 | - | 9e-21 | 24.17% (480) | ABC transporter |
| M. abscessus ATCC 19977 | MAB_2803c | - | 6e-29 | 35.24% (227) | methionine import ATP-binding protein metN |
| M. marinum M | MMAR_5151 | dppD | 9e-28 | 27.55% (530) | dipeptide-transport ATP-binding protein ABC transporter DppD |
| M. avium 104 | MAV_3418 | - | 6e-25 | 25.68% (514) | ABC transporter, ATP-binding protein DppD |
| M. thermoresistible (build 8) | TH_1784 | - | 5e-29 | 33.04% (224) | ABC transporter, ATP-binding protein GluA |
| M. ulcerans Agy99 | MUL_4237 | dppD | 1e-27 | 27.55% (530) | dipeptide-transport ATP-binding protein ABC transporter DppD |
| M. vanbaalenii PYR-1 | Mvan_4009 | - | 9e-36 | 36.76% (253) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1784|M.thermoresistible__bu --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 --------------------------------------------------
Rv3663c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5151|M.marinum_M --------------------------------------------------
MUL_4237|M.ulcerans_Agy99 --------------------------------------------------
MAV_3418|M.avium_104 --------------------------------------------------
MLBr_00848|M.leprae_Br4923 MDLCIHRRMALLCQATRRCHRWKTQLGRCAAPDLAPGTGTIGKVTAHDPR
Mflv_2607|M.gilvum_PYR-GCK ---MTISVEKASSDSKPGGKVPLVELKNIGKSYGN----ITALKDISLRV
Mvan_4009|M.vanbaalenii_PYR-1 ---MTISVEKPSSDAQAGGKVPLVELKNVGKSYGN----ITALKDICLRV
MSMEG_4000|M.smegmatis_MC2_155 ------------MSASRSLAAPLLKVDAVTKRYIG----VTALTDASLAV
MAB_2803c|M.abscessus_ATCC_199 ----------------MGDVAPLIQFQNVTKTFRVGKKTVTAVDDVSLDI
TH_1784|M.thermoresistible__bu ---MDSAGADVAVADPAAEAVPMISMRGVNKYFGS----LHVLRDINLDV
Mb3687c|M.bovis_AF2122/97 ---------------MSVPAAPLLSVEGLEVTFGTDAPAVCG---VDLAV
Rv3663c|M.tuberculosis_H37Rv ---------------MSVPAAPLLSVEGLEVTFGTDAPAVCG---VDLAV
MMAR_5151|M.marinum_M ---------------MSTAAAPLLSVHGLQVRFGTDEPVVAG---MDLSV
MUL_4237|M.ulcerans_Agy99 ---------------MSTAAAPLLSVHGLQVRFGTDEPVVAG---MDLSV
MAV_3418|M.avium_104 ------------------MGPPLLDVSGLTVTYPGAVPRVALD-RVDLAV
MLBr_00848|M.leprae_Br4923 HLHHRSGSLQPGELAQASVLAALCAVTAIVSVVVPFAAGLALLGTVPMGL
.: . : : : :
Mflv_2607|M.gilvum_PYR-GCK HA-------------------------------------GEVTGILGDNG
Mvan_4009|M.vanbaalenii_PYR-1 HA-------------------------------------GEVTGILGDNG
MSMEG_4000|M.smegmatis_MC2_155 GA-------------------------------------GEVRALLGRNG
MAB_2803c|M.abscessus_ATCC_199 GS-------------------------------------GDVFAMIGYSG
TH_1784|M.thermoresistible__bu AR-------------------------------------GQVVVVLGPSG
Mb3687c|M.bovis_AF2122/97 RS-------------------------------------GQTVAVVGESG
Rv3663c|M.tuberculosis_H37Rv RS-------------------------------------GQTVAVVGESG
MMAR_5151|M.marinum_M YR-------------------------------------GQTVAVVGESG
MUL_4237|M.ulcerans_Agy99 YR-------------------------------------GQTVAVVGESG
MAV_3418|M.avium_104 SS-------------------------------------GEFVAVVGESG
MLBr_00848|M.leprae_Br4923 LAYRYRFRVLMTAMVAAGVIAFLITGLGGFIAVVNSAYIGGLTGVVKRKG
* :: .*
Mflv_2607|M.gilvum_PYR-GCK AGKSTLIKIIAGLHQ----QTDGELLVDG---------------------
Mvan_4009|M.vanbaalenii_PYR-1 AGKSTLIKIIAGLHQ----QTEGQLLVDG---------------------
MSMEG_4000|M.smegmatis_MC2_155 AGKSTLIRILSGVEQ----ADTGTVSIDG---------------------
MAB_2803c|M.abscessus_ATCC_199 AGKSTLVRLINGLER----PTSGRVIVDG---------------------
TH_1784|M.thermoresistible__bu SGKSTLCRTLNRLEP----IDSGAISIDG---------------------
Mb3687c|M.bovis_AF2122/97 SGKSTTAAAILGLLPAGGRITAGRVVFDG---------------------
Rv3663c|M.tuberculosis_H37Rv SGKSTTAAAILGLLPAGGRITAGRVVFDG---------------------
MMAR_5151|M.marinum_M SGKSTAAAAILGLLAPGGRITAGRIIFDG---------------------
MUL_4237|M.ulcerans_Agy99 SGKSTAAAAILGLLAPGGRITAGRIIFDG---------------------
MAV_3418|M.avium_104 SGKTTLANAVVGLLPAAARITAGSLTIAG---------------------
MLBr_00848|M.leprae_Br4923 QGTLTVIALALFAGLAFGAANVVALVVLGRLRHLIFKAMTANVDGIAATL
*. * : . *
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1784|M.thermoresistible__bu --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 --------------------------------------------------
Rv3663c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5151|M.marinum_M --------------------------------------------------
MUL_4237|M.ulcerans_Agy99 --------------------------------------------------
MAV_3418|M.avium_104 --------------------------------------------------
MLBr_00848|M.leprae_Br4923 TWMHLPWVAVQLKRYFADGLQHWPWMLLGYFVITILVVSLIGWWVLSRVL
Mflv_2607|M.gilvum_PYR-GCK ---------------EPTRFASPADALGKG---IATVYQNLAVVPLMPVW
Mvan_4009|M.vanbaalenii_PYR-1 ---------------EPTKFASPAEALGKG---IATVYQNLAVVPLMPVW
MSMEG_4000|M.smegmatis_MC2_155 ---------------TILDGGGVRRANELG---VQTVHQELSLVPDMTVA
MAB_2803c|M.abscessus_ATCC_199 ---------------AEVSALGERELRGLR-SDIGMIFQQFNLFRSRTVA
TH_1784|M.thermoresistible__bu ---------------VALPAEG-RELARLR-ADVGMVFQQFNLFAHKTIV
Mb3687c|M.bovis_AF2122/97 ---------------RDITGADAKRLRSIRGREIGYVPQDPMTNLNPVWK
Rv3663c|M.tuberculosis_H37Rv ---------------RDITGADAKRLRSIRGREIGYVPQDPMTNLNPVWK
MMAR_5151|M.marinum_M ---------------RDITSADRRVMRTIRGREIGYVPQDPMTNLNPVWK
MUL_4237|M.ulcerans_Agy99 ---------------RDITSADRRVMRTIRGREIGYVPQDPMTNLNPVWK
MAV_3418|M.avium_104 ---------------RDVLGLSERQWCRLRGSTVGLVPQDPATSLNPVRS
MLBr_00848|M.leprae_Br4923 ERIRDIPDVHKLDAPSACNEDAPVGPVPVWLDKVRFRYPHAGQDALREVS
: .
Mflv_2607|M.gilvum_PYR-GCK RNFFLGQELRKKRFP----------FSLDANAMRATTLTELAKMGID---
Mvan_4009|M.vanbaalenii_PYR-1 RNFFLGQELRKKSFP----------FSLDADAMRATTLTELSKMGID---
MSMEG_4000|M.smegmatis_MC2_155 ENMFVGAWPRSGGK-------------IDYDAMRAEASTVIDRLGLD---
MAB_2803c|M.abscessus_ATCC_199 GNIAY-PLRVAGWT-------------ADKRNARVAELLEFVGLGD----
TH_1784|M.thermoresistible__bu DNVALGPMKVRKTP-------------REQARAEAMALLERVGVAD----
Mb3687c|M.bovis_AF2122/97 VGFQVTEALRANTD-------------GRAARRRAVELLAEAGLPDP---
Rv3663c|M.tuberculosis_H37Rv VGFQVTEALRANTD-------------GRAARRRAVELLAEAGLPDP---
MMAR_5151|M.marinum_M VGFQIREALRANTD-------------GRDARRRTVELLAQAGMPDP---
MUL_4237|M.ulcerans_Agy99 VGFQIREALRANTD-------------GRDARRRTVELLAQAGMPDP---
MAV_3418|M.avium_104 IGSQIAEIFPLKGER----------LSRREARRRCIELLDRVEIDRP---
MLBr_00848|M.leprae_Br4923 LDLRVGEHVAVTGANGSGKTTLMLILAGREPTSGTVDRPGAVGLGKLGGT
. :
Mflv_2607|M.gilvum_PYR-GCK -----------------------------------------LPDVDVPIG
Mvan_4009|M.vanbaalenii_PYR-1 -----------------------------------------LPDVDVPIG
MSMEG_4000|M.smegmatis_MC2_155 -----------------------------------------I-DPDSPVA
MAB_2803c|M.abscessus_ATCC_199 -------------------------------------------KAKSYPD
TH_1784|M.thermoresistible__bu -------------------------------------------QADKYPA
Mb3687c|M.bovis_AF2122/97 -----------------------------------------AKQAGRYPH
Rv3663c|M.tuberculosis_H37Rv -----------------------------------------AKQAGRYPH
MMAR_5151|M.marinum_M -----------------------------------------DKHAQRYPH
MUL_4237|M.ulcerans_Agy99 -----------------------------------------DKHAQRYPH
MAV_3418|M.avium_104 -----------------------------------------AQRLKQYPG
MLBr_00848|M.leprae_Br4923 AVVLQHPESQVLGTRVADDVVWGLPPGTDVDVNRLLREVGLDAFAERDTG
Mflv_2607|M.gilvum_PYR-GCK SLSGGQKQCVAIARAVFFGARVLILDEPTAALGVKQSGVVLKYIT-AAKE
Mvan_4009|M.vanbaalenii_PYR-1 SLSGGQKQCVAIARAVFFGARVLILDEPTAALGVKQSGVVLKYIT-AAKE
MSMEG_4000|M.smegmatis_MC2_155 ALSLATRQLVEICRAVRRQPRVLILDEPTSALAATEVSIVLDTVT-RIAQ
MAB_2803c|M.abscessus_ATCC_199 QLSGGQKQRVGIARALATSPSVLLADEATSALDPETTSDVLRLLREVNAE
TH_1784|M.thermoresistible__bu QLSGGQQQRVAIARSLAMNPKVMLFDEPTSALDPEMINEVLAVMSALAAD
Mb3687c|M.bovis_AF2122/97 QLSGGMCQRALIAIGLAGRPRLLIADEPTSALDVTVQRQVLDHLQGLTDE
Rv3663c|M.tuberculosis_H37Rv QLSGGMCQRALIAIGLAGRPRLLIADEPTSALDVTVQRQVLDHLQGLTDE
MMAR_5151|M.marinum_M QLSGGMCQRALIAIGLAGRPALLIADEPTSALDVTVQRQVLDHLQRRTDE
MUL_4237|M.ulcerans_Agy99 QLSGGMCQRALIAIGLAGRPALLIADEPTSALDVTVQRQVLDHLQRRTDE
MAV_3418|M.avium_104 ELSGGMRQRVLIAIAFALNPGLLIADEPTSALDVTVQRHVLAVFDRLVAE
MLBr_00848|M.leprae_Br4923 SLSGGELQRLALAAALAREPSLLIADEVTSMVDRQGRDALLGVLSGLTKR
** . * :. .. . ::: ** *: : :* .
Mflv_2607|M.gilvum_PYR-GCK AGFGVVFITHNPHHAHLVGDHFVLLNRGRQKLDCSYDDITLEHLTQQMAG
Mvan_4009|M.vanbaalenii_PYR-1 AGFGVVFITHNPHHAHLVGDHFVLLNRGRQKLDCTYDDITLEHLTQEMAG
MSMEG_4000|M.smegmatis_MC2_155 TGVAVIYVSHRLDEIRRIASSATILRDGRVVDTVEMDTTGTDAMIAMMVG
MAB_2803c|M.abscessus_ATCC_199 FGVTIVVITHEMDVVRTVADHVAVLADGKVVETGTVFDVFSTPQSEAGKR
TH_1784|M.thermoresistible__bu -GMTMLVVTHEMGFARRASDRVVFMSDGAIVEDAEPARFFEDPQTDRARD
Mb3687c|M.bovis_AF2122/97 LGTALLLITHDLALAAQRAEAVVVVRRGVVVESGAAQSILQSPQHEYTRR
Rv3663c|M.tuberculosis_H37Rv LGTALLLITHDLALAAQRAEAVVVVRRGVVVESGAAQSILQSPQHEYTRR
MMAR_5151|M.marinum_M LGTALLLITHDLALAAERAESVLVVHHGVVVESGTASDILRSPQHEYTKR
MUL_4237|M.ulcerans_Agy99 VGTALLLITHDLALAAERAESVLVVDHGVVVESGTASDILRSPQHEYTKR
MAV_3418|M.avium_104 QRTTVVFITHDIGVATDHASRIVVMRGGVVVEDAPAEAIVAAPRTEYAAH
MLBr_00848|M.leprae_Br4923 HPIALVHITHYNNEADTADRTINLSDS---PDNAGMAETVAPPVSTVAVD
:: ::* .
Mflv_2607|M.gilvum_PYR-GCK GDELEALSHELRAAKQ----------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 GDELEALTHELRAAKS----------------------------------
MSMEG_4000|M.smegmatis_MC2_155 PDHQAQERPPLRAVDRTRTPLLSVRDLRLPP-------------------
MAB_2803c|M.abscessus_ATCC_199 FVG-----------------------------------------------
TH_1784|M.thermoresistible__bu FLG-----------------------------------------------
Mb3687c|M.bovis_AF2122/97 LVAAAPSLTARSRRPPESR----SRATTQAG---DILVVSELTKIYRESR
Rv3663c|M.tuberculosis_H37Rv LVAAAPSLTARSRRPPESR----SRATTQAG---DILVVSELTKIYRESR
MMAR_5151|M.marinum_M LVAAAPSLTVRDPRRAPARGPVPGGADQPAGGCGDILSVSELTKIYRESR
MUL_4237|M.ulcerans_Agy99 LVAAAPSLTVRDPRRAPARGPVPGGADQPAGGCGDILSVSELTKIYRESR
MAV_3418|M.avium_104 LVR---RLTAPAP------------AAAPAG-SEDVIEVAGVAKEFRLG-
MLBr_00848|M.leprae_Br4923 HRPHAPVLELVGVGHEYGSG------------------------------
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 ----------KIEKVSFDLFAGEVLGIAGLMGSGRTEVLRAVAGFDTPVS
MAB_2803c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1784|M.thermoresistible__bu --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 GAPWRRVESRAVDGVSFRLPRASTLAIVGESGSGKSTLARMVLGLLQPTS
Rv3663c|M.tuberculosis_H37Rv GAPWRRVESRAVDGVSFRLPRASTLAIVGESGSGKSTLARMVLGLLQPTS
MMAR_5151|M.marinum_M GAPWRRGEFRAVDSVSFRLRRASTLAIVGESGSGKSTVARMVLGLLQPTS
MUL_4237|M.ulcerans_Agy99 GAPWRRGEFRAVDSVSFRLRRASTLAIVGESGSGKSTVARMVLGLLQPTS
MAV_3418|M.avium_104 ----ARARHTAVAEVAFTVRRGETLALVGESGSGKSTTAKMVVGLLTPTR
MLBr_00848|M.leprae_Br4923 ----TPWAKAALHDISFVVRQGDGVLVYGSNGSGKSTLAWIMAGLMVPTT
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 GEMTLDGKPMPWNAPAVMKRNGVGLTPEDRKHDAIVPLLGVDENMVLSDY
MAB_2803c|M.abscessus_ATCC_199 ------------------------------------------TVLRNTPS
TH_1784|M.thermoresistible__bu ------------------------------------------KILHH---
Mb3687c|M.bovis_AF2122/97 GTVVFDGT-YDVGALARDQVLAFRRRVQPVFQNPYSSLDPMYSVFRAIEE
Rv3663c|M.tuberculosis_H37Rv GTVVFDGT-YDVGALARDQVLAFRRRVQPVFQNPYSSLDPMYSVFRAIEE
MMAR_5151|M.marinum_M GTVSFDGARIQPDKLDRDAQLAFRRRIQPVFQNPYSSLDPMYSVFRAIEE
MUL_4237|M.ulcerans_Agy99 GTVSFDGARIQPDKLDRDAQLAFRCRIQPVFQNPYSSLDPMYSVFRAIEE
MAV_3418|M.avium_104 GTVRVLGR--DVAALPARARRRHWQDIQFVYQNPGSALDPRWTVRQILDG
MLBr_00848|M.leprae_Br4923 GACLIDGR-----------------PTHEHVGAVALSFQAARLQLMRSRV
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 TRVRHHGPISLRRMRTAAQSLIDRLRIETASPVTPIVNLSGGNQQKAIIG
MAB_2803c|M.abscessus_ATCC_199 G-EDIARIAQRHKGRIVAVEISEGVQIG------------------PALA
TH_1784|M.thermoresistible__bu --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 P-LRVHHVGDRRQRQRAVRELVDQVALPSSILGRRPRELSGGQRQRVAIA
Rv3663c|M.tuberculosis_H37Rv P-LRVHHVGDRRQRQRAVRELVDQVALPSSILGRRPRELSGGQRQRVAIA
MMAR_5151|M.marinum_M P-LRIHRVGDRAQRRQMVAELVDQVALPSSVLGRLPRELSGGQRQRVAIA
MUL_4237|M.ulcerans_Agy99 P-LRIHRVGDRGQRRQMVAELVDQVALPSSVLGRLPRELSGGQRQRVAIA
MAV_3418|M.avium_104 P-LRSYRVDDRARRAQRIAEALASVGLSDDKLDRRPGELSGGECQRVAIA
MLBr_00848|M.leprae_Br4923 DLEVASAAGFSPRDEDRVAAALGVVGLDPALAKRRIDQLSGGQMRRVVLA
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 RWLHADASILLLDEPTRGVDVAAKAGIYDLVR-ALADAGKAVIFVSGELE
MAB_2803c|M.abscessus_ATCC_199 RATELGVRFEVVYGGITTLQSRSFGNLTLELEGPDDQVAHVVSDLSAVTT
TH_1784|M.thermoresistible__bu --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 RALALRPEVLVCDEAVSALDVLVQAQILDLLADLQADLGLTYLFISHDLA
Rv3663c|M.tuberculosis_H37Rv RALALRPEVLVCDEAVSALDVLVQAQILDLLADLQADLGLTYLFISHDLA
MMAR_5151|M.marinum_M RALALRPDVLVCDEAVSALDVLVQAQILDLLAELQADLGLAYLFISHDLA
MUL_4237|M.ulcerans_Agy99 RALALRPDVLVCDEAVSALDVLVQAQILDLLAELQADLGLAYLFISHDLA
MAV_3418|M.avium_104 RALVLRPQIVVLDEPLSALDVVTQDQIVGLLLRLQRQLGLTYLFVSHDLS
MLBr_00848|M.leprae_Br4923 GLLACSPRALILDEPLAGLDAVSQRGLLRLLEDLRCERGLTVVVTSHDFV
Mflv_2607|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4009|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4000|M.smegmatis_MC2_155 ELPLVCDRVITVQGGRVTAEFIGVDVTVDAVLSAAMAA------------
MAB_2803c|M.abscessus_ATCC_199 VQRVAR--------------------------------------------
TH_1784|M.thermoresistible__bu --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 VIRQIADDVLVMRAGRVVEHASTEEVFSRPRHEYTRQLLQAIPGAPSAPR
Rv3663c|M.tuberculosis_H37Rv VIRQIADDVLVMRAGRVVEHASTEEVFSRPRHEYTRQLLQAIPGAPSAPR
MMAR_5151|M.marinum_M VVRQISDDVLVMRAGQVVEHAPTEEVFTRPGAQYTQRLLEAIPGGPVAPR
MUL_4237|M.ulcerans_Agy99 VVRQISDDVLVMRAGQVVEHAPTEEVFTRPGAQYTQRLLEAIPGGPVAPR
MAV_3418|M.avium_104 VVRRLAHRVVVLRAGRVVETGDTAAVFAAPAADYTRALLDAVPGRRLAHP
MLBr_00848|M.leprae_Br4923 GLEDVCPRTVHLRNGALESVSTTAGGTS----------------------
Mflv_2607|M.gilvum_PYR-GCK -----
Mvan_4009|M.vanbaalenii_PYR-1 -----
MSMEG_4000|M.smegmatis_MC2_155 -----
MAB_2803c|M.abscessus_ATCC_199 -----
TH_1784|M.thermoresistible__bu -----
Mb3687c|M.bovis_AF2122/97 KVGNL
Rv3663c|M.tuberculosis_H37Rv KVGNL
MMAR_5151|M.marinum_M -----
MUL_4237|M.ulcerans_Agy99 -----
MAV_3418|M.avium_104 ALSPS
MLBr_00848|M.leprae_Br4923 -----