For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GPPLLDVSGLTVTYPGAVPRVALDRVDLAVSSGEFVAVVGESGSGKTTLANAVVGLLPAAARITAGSLTI AGRDVLGLSERQWCRLRGSTVGLVPQDPATSLNPVRSIGSQIAEIFPLKGERLSRREARRRCIELLDRVE IDRPAQRLKQYPGELSGGMRQRVLIAIAFALNPGLLIADEPTSALDVTVQRHVLAVFDRLVAEQRTTVVF ITHDIGVATDHASRIVVMRGGVVVEDAPAEAIVAAPRTEYAAHLVRRLTAPAPAAAPAGSEDVIEVAGVA KEFRLGARARHTAVAEVAFTVRRGETLALVGESGSGKSTTAKMVVGLLTPTRGTVRVLGRDVAALPARAR RRHWQDIQFVYQNPGSALDPRWTVRQILDGPLRSYRVDDRARRAQRIAEALASVGLSDDKLDRRPGELSG GECQRVAIARALVLRPQIVVLDEPLSALDVVTQDQIVGLLLRLQRQLGLTYLFVSHDLSVVRRLAHRVVV LRAGRVVETGDTAAVFAAPAADYTRALLDAVPGRRLAHPALSPS*
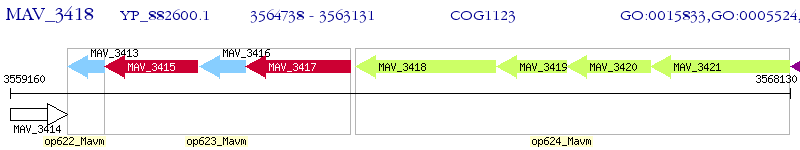
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3418 | - | - | 100% (535) | ABC transporter, ATP-binding protein DppD |
| M. avium 104 | MAV_0467 | dppD | e-122 | 47.11% (554) | ABC transporter, ATP-binding protein DppD |
| M. avium 104 | MAV_1431 | oppD | 3e-84 | 37.85% (605) | ABC transporter, ATP-binding protein OppD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3687c | dppD | 1e-124 | 49.26% (540) | peptide ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_2758 | - | 1e-102 | 43.05% (532) | AAA ATPase |
| M. tuberculosis H37Rv | Rv3663c | dppD | 1e-124 | 49.26% (540) | peptide ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_01122 | - | 2e-56 | 47.17% (265) | putative ABC transport protein, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_0429 | - | 1e-108 | 43.90% (533) | putative oligopeptide ABC transporter, ATP-binding protein |
| M. marinum M | MMAR_5151 | dppD | 1e-121 | 47.99% (548) | dipeptide-transport ATP-binding protein ABC transporter DppD |
| M. smegmatis MC2 155 | MSMEG_2844 | - | 0.0 | 68.77% (538) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4070 | - | 1e-108 | 43.88% (531) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_4237 | dppD | 1e-120 | 47.81% (548) | dipeptide-transport ATP-binding protein ABC transporter DppD |
| M. vanbaalenii PYR-1 | Mvan_3776 | - | 1e-107 | 44.44% (513) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3418|M.avium_104 ----MGPPLLDVSGLTVTYPG---AVPRVALDRVDLAVSSGEFVAVVGES
MSMEG_2844|M.smegmatis_MC2_155 ------MPLLELSDLTVTYPGDG-ITPKIALGHVNLSLAAGEFAAIVGES
Mb3687c|M.bovis_AF2122/97 -MSVPAAPLLSVEGLEVTFGTD---APAVCG--VDLAVRSGQTVAVVGES
Rv3663c|M.tuberculosis_H37Rv -MSVPAAPLLSVEGLEVTFGTD---APAVCG--VDLAVRSGQTVAVVGES
MMAR_5151|M.marinum_M -MSTAAAPLLSVHGLQVRFGTD---EPVVAG--MDLSVYRGQTVAVVGES
MUL_4237|M.ulcerans_Agy99 -MSTAAAPLLSVHGLQVRFGTD---EPVVAG--MDLSVYRGQTVAVVGES
MLBr_01122|M.leprae_Br4923 -----MSPLLEVTDLAVTFETN--GEPVTAVRGISYRINAGEVVAMVGES
Mflv_2758|M.gilvum_PYR-GCK ------MTVLNVQDLSVRIGAR----TIVDS--VSFAVEREKTVGIVGES
Mvan_3776|M.vanbaalenii_PYR-1 ------MTVLSVDNLRVRIGAR----TIVDG--VSFDVAREKTVGIVGES
TH_4070|M.thermoresistible__bu ---VVTGPVLDVRDLRVRIGRR----EIVRG--VSFAVARGQTLGIVGES
MAB_0429|M.abscessus_ATCC_1997 MEVPTSDSVVDITGLTVDFEVDKQWVPAVKG--VSLSVAKGEVLAIVGES
.::.: .* * :. : : .:****
MAV_3418|M.avium_104 GSGKTTLANAVVGLLPAAARITAGSLTIAG----RDVLGLSERQWCRLRG
MSMEG_2844|M.smegmatis_MC2_155 GSGKTTLVNTVLGLLPRTATVTADTLALDG----AGILGLTEREWSRIRG
Mb3687c|M.bovis_AF2122/97 GSGKSTTAAAILGLLPAGGRITAGRVVFDG----RDITGADAKRLRSIRG
Rv3663c|M.tuberculosis_H37Rv GSGKSTTAAAILGLLPAGGRITAGRVVFDG----RDITGADAKRLRSIRG
MMAR_5151|M.marinum_M GSGKSTAAAAILGLLAPGGRITAGRIIFDG----RDITSADRRVMRTIRG
MUL_4237|M.ulcerans_Agy99 GSGKSTAAAAILGLLAPGGRITAGRIIFDG----RDITSADRRVMRTIRG
MLBr_01122|M.leprae_Br4923 GSGKSAAAMAVVGLLPEYAQVRG-SVRLHN----TELVGLADKTMSRFRG
Mflv_2758|M.gilvum_PYR-GCK GSGKTMTVLAATGLLDAPGARVSGVSTLGG---EVHLVGATPKALRSVHG
Mvan_3776|M.vanbaalenii_PYR-1 GSGKTMTVLAATGLLDAPGARVSGSSTLTGADEPVQLVGASPRVLRVVHG
TH_4070|M.thermoresistible__bu GSGKSMTVLAAAGLLDAPGAVVTGSATVGG----VELIGAPPRLLRSVHG
MAB_0429|M.abscessus_ATCC_1997 GSGKSTTAMAIPALLP-PSARVRGSVKLNG----AELLGATNDELRAVRG
****: . : .** . . . : . .:*
MAV_3418|M.avium_104 STVGLVPQDPATSLNPVRSIGSQIAEIFPLKGERLSRREARRRCIELLDR
MSMEG_2844|M.smegmatis_MC2_155 TTVGLVPQDPGASLNPVRTIGSQIAEIFDLRDEKLSRAERRRRCIDLLEQ
Mb3687c|M.bovis_AF2122/97 REIGYVPQDPMTNLNPVWKVGFQVTEALRANTD---GRAARRRAVELLAE
Rv3663c|M.tuberculosis_H37Rv REIGYVPQDPMTNLNPVWKVGFQVTEALRANTD---GRAARRRAVELLAE
MMAR_5151|M.marinum_M REIGYVPQDPMTNLNPVWKVGFQIREALRANTD---GRDARRRTVELLAQ
MUL_4237|M.ulcerans_Agy99 REIGYVPQDPMTNLNPVWKVGFQIREALRANTD---GRDARRRTVELLAQ
MLBr_01122|M.leprae_Br4923 KAIGTVFQDPMSALTPVYTVGDQITEAIKVHQPRVGKKAAHRRAVELLEL
Mflv_2758|M.gilvum_PYR-GCK ARIGFVFQDPGTSLNPLLTLERQITESLETHRN-LTRRAANDRALELLEA
Mvan_3776|M.vanbaalenii_PYR-1 ARIGFVFQDPGTSLNPLLTLERQITESLEKHRH-LTRRAAKDRALELLEA
TH_4070|M.thermoresistible__bu NRVGFVFQDPGTSLNPLLTLERQITEVLEAHRG-MTRRAARGRALELLTA
MAB_0429|M.abscessus_ATCC_1997 KDVAVIFQEPMTALNPVYTIGWQIAEAVCAHKK-MSRRQARDRAVELLDL
:. : *:* : *.*: .: *: * . . . * ::**
MAV_3418|M.avium_104 VEIDRPAQRLKQYPGELSGGMRQRVLIAIAFALNPGLLIADEPTSALDVT
MSMEG_2844|M.smegmatis_MC2_155 VEIDQPERRLRQHPGELSGGMRQRVLIAIAFGLNPKLLVADEPTSALDVT
Mb3687c|M.bovis_AF2122/97 AGLPDPAKQAGRYPHQLSGGMCQRALIAIGLAGRPRLLIADEPTSALDVT
Rv3663c|M.tuberculosis_H37Rv AGLPDPAKQAGRYPHQLSGGMCQRALIAIGLAGRPRLLIADEPTSALDVT
MMAR_5151|M.marinum_M AGMPDPDKHAQRYPHQLSGGMCQRALIAIGLAGRPALLIADEPTSALDVT
MUL_4237|M.ulcerans_Agy99 AGMPDPDKHAQRYPHQLSGGMCQRALIAIGLAGRPALLIADEPTSALDVT
MLBr_01122|M.leprae_Br4923 VGITQPDRRARAFPHELSGGERQRVVIATAIANDPDLLICDEPTTALDVT
Mflv_2758|M.gilvum_PYR-GCK VGLPEPQMRLHAYPHQLSGGQRQRVMIAIALACDPELLIADEPTTALDVT
Mvan_3776|M.vanbaalenii_PYR-1 VGLPEPQMRLHAYPHQLSGGQRQRVMIAIALACDPELLVADEPTTALDVT
TH_4070|M.thermoresistible__bu VGLPDPAARLRSYPHQLSGGQRQRVMIAIALACDPEVLIADEPTTALDVT
MAB_0429|M.abscessus_ATCC_1997 VDMPEPAKRVKHYPHQLSGGQRQRAMIAQALALDPGLLIADEPTTALDVT
. : * : .* :**** **.:** .:. * :*:.****:*****
MAV_3418|M.avium_104 VQRHVLAVFDRLVAEQRTTVVFITHDIGVATDHASRIVVMRGGVVVEDAP
MSMEG_2844|M.smegmatis_MC2_155 VQAQVLRVFDRLVAEHGTTVLFVTHDIGVATDHASRVVVMRKGQIVEDAP
Mb3687c|M.bovis_AF2122/97 VQRQVLDHLQGLTDELGTALLLITHDLALAAQRAEAVVVVRRGVVVESGA
Rv3663c|M.tuberculosis_H37Rv VQRQVLDHLQGLTDELGTALLLITHDLALAAQRAEAVVVVRRGVVVESGA
MMAR_5151|M.marinum_M VQRQVLDHLQRRTDELGTALLLITHDLALAAERAESVLVVHHGVVVESGT
MUL_4237|M.ulcerans_Agy99 VQRQVLDHLQRRTDEVGTALLLITHDLALAAERAESVLVVDHGVVVESGT
MLBr_01122|M.leprae_Br4923 VQAQILEVLKTARDVTGAGVLIITHDLGVVTEFADQALVMYAGRVVESAQ
Mflv_2758|M.gilvum_PYR-GCK TQAQIIELVRTLQRDFGTAVVWISHDLGVIGQVADDVTVMRNGIAVEQAP
Mvan_3776|M.vanbaalenii_PYR-1 TQSQIIELVRELQRDFGTAVVWISHDLGVIGQVADEVTVMRNGVTVEQAP
TH_4070|M.thermoresistible__bu TQAQIIELVRELQRDFGTAVVWISHDLGVIGEVADEVTVLRHGAAVEQAH
MAB_0429|M.abscessus_ATCC_1997 VQAEILDLMRDLRHRIDAGIILITHDMGVVADMADRIMVMKDGEVVEQGD
.* .:: . : :: ::**:.: : *. *: * **..
MAV_3418|M.avium_104 AEAIVAAPRTEYAA-------HLVRRLTA---PAPAAAP-----------
MSMEG_2844|M.smegmatis_MC2_155 VDDIVSRPSSEYTA-------ALVRRIGAGITPPAAAHENV---------
Mb3687c|M.bovis_AF2122/97 AQSILQSPQHEYTR-------RLVAAAPSLTARSRRPPESR---------
Rv3663c|M.tuberculosis_H37Rv AQSILQSPQHEYTR-------RLVAAAPSLTARSRRPPESR---------
MMAR_5151|M.marinum_M ASDILRSPQHEYTK-------RLVAAAPSLTVRDPRRAPARG--------
MUL_4237|M.ulcerans_Agy99 ASDILRSPQHEYTK-------RLVAAAPSLTVRDPRRAPARG--------
MLBr_01122|M.leprae_Br4923 VSTLYRERQMPYTVGLLGSVPRLDATQGTRLVPIPGAPPSLAGLQQECPF
Mflv_2758|M.gilvum_PYR-GCK IMAVFDNPRDDYTR-------DLLAARPVLERAGPGPVTDAE--------
Mvan_3776|M.vanbaalenii_PYR-1 IMEVFDRPREEYTR-------GLLAARPVLDRAGPPAVADAE--------
TH_4070|M.thermoresistible__bu IAEVFERPRHPYTR-------ELLHARPRLGVGGPPQVTSTE--------
MAB_0429|M.abscessus_ATCC_1997 AEGIFHRAQQPYTR-------QLLASVPHLGATLHGRDESNP--------
: *: *
MAV_3418|M.avium_104 --------------------------------------------------
MSMEG_2844|M.smegmatis_MC2_155 --------------------------------------------------
Mb3687c|M.bovis_AF2122/97 ------------------------------------------------SR
Rv3663c|M.tuberculosis_H37Rv ------------------------------------------------SR
MMAR_5151|M.marinum_M ---------------------------------------------PVPGG
MUL_4237|M.ulcerans_Agy99 ---------------------------------------------PVPGG
MLBr_01122|M.leprae_Br4923 APRCPLVVDECRLREPELVEVRAGHRTACIRTDQVSGRSAADIYGINTDT
Mflv_2758|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3776|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4070|M.thermoresistible__bu --------------------------------------------------
MAB_0429|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3418|M.avium_104 --AGSE---DVIEVAGVAKEFRLG-----ARAR--HTAVAEVAFTVRRGE
MSMEG_2844|M.smegmatis_MC2_155 -HAPAD---EVIRVAHVSKEFRLD-----RRHR--HRAVDDISFTVRRGE
Mb3687c|M.bovis_AF2122/97 ATTQAG---DILVVSELTKIYRESRGAPWRRVE--SRAVDGVSFRLPRAS
Rv3663c|M.tuberculosis_H37Rv ATTQAG---DILVVSELTKIYRESRGAPWRRVE--SRAVDGVSFRLPRAS
MMAR_5151|M.marinum_M ADQPAGGCGDILSVSELTKIYRESRGAPWRRGE--FRAVDSVSFRLRRAS
MUL_4237|M.ulcerans_Agy99 ADQPAGGCGDILSVSELTKIYRESRGAPWRRGE--FRAVDSVSFRLRRAS
MLBr_01122|M.leprae_Br4923 RTSATSDSSVVVRVRDLVKTYRLTKGVGLRRAIGEIRAVNGVSFELQQGR
Mflv_2758|M.gilvum_PYR-GCK ---------TLLDVDGLDVSFAVSSPV--GRS--TVHAVKDVSFRIRRGT
Mvan_3776|M.vanbaalenii_PYR-1 ---------RLLEVDGLDVRFAVTTPV--GRS--TVHAVRDVSFTVRRGT
TH_4070|M.thermoresistible__bu ---------VLLRVEGLQVRYDVSTPT--GRS--TVRAVSDLSLSIRRGT
MAB_0429|M.abscessus_ATCC_1997 ---------PPTDLA-LSVEHAVIEYP--GKSG-SFRAVDDVSFTIGRGE
: : . : ** ::: : :.
MAV_3418|M.avium_104 TLALVGESGSGKSTTAKMVVGLLTPTRGTVRVLGR--DVAALPARARRRH
MSMEG_2844|M.smegmatis_MC2_155 TLALVGESGSGKSTTAKMIIGLTQPTRGSIEVLGH--DITRLGHRDRRAH
Mb3687c|M.bovis_AF2122/97 TLAIVGESGSGKSTLARMVLGLLQPTSGTVVFDGT-YDVGALARDQVLAF
Rv3663c|M.tuberculosis_H37Rv TLAIVGESGSGKSTLARMVLGLLQPTSGTVVFDGT-YDVGALARDQVLAF
MMAR_5151|M.marinum_M TLAIVGESGSGKSTVARMVLGLLQPTSGTVSFDGARIQPDKLDRDAQLAF
MUL_4237|M.ulcerans_Agy99 TLAIVGESGSGKSTVARMVLGLLQPTSGTVSFDGARIQPDKLDRDAQLAF
MLBr_01122|M.leprae_Br4923 TLGIVGESGSGKSTTLHEILRLAVPQSGSIEVLGI--DVATLNRASRRSM
Mflv_2758|M.gilvum_PYR-GCK TLGLVGESGSGKSTVAAALTGLVTPDAGTATLAGD--DVFSVKGRAARDM
Mvan_3776|M.vanbaalenii_PYR-1 TLGLVGESGSGKSTVAACLTGLVKPDAGTANLAGV--DVLHVKGSAAKRI
TH_4070|M.thermoresistible__bu TLGLVGESGSGKSTVAAALTGLVRPTAGRATLAGV--DLFGVRGRDRRRL
MAB_0429|M.abscessus_ATCC_1997 VVGLVGESGSGKSTIGRAAVGLLRVASGTICVAGQ--DISTANRKQLRDI
.:.:********** * * . * :
MAV_3418|M.avium_104 WQDIQFVYQNPGSALDPRWTVRQILDGPLRSYRVDD-RARRAQRIAEALA
MSMEG_2844|M.smegmatis_MC2_155 WKNIQFVYQNPDSALDPRWSVREVLESPLRAYHLDN-RD---QRIAEALD
Mb3687c|M.bovis_AF2122/97 RRRVQPVFQNPYSSLDPMYSVFRAIEEPLRVHHVGD-RRQRQRAVRELVD
Rv3663c|M.tuberculosis_H37Rv RRRVQPVFQNPYSSLDPMYSVFRAIEEPLRVHHVGD-RRQRQRAVRELVD
MMAR_5151|M.marinum_M RRRIQPVFQNPYSSLDPMYSVFRAIEEPLRIHRVGD-RAQRRQMVAELVD
MUL_4237|M.ulcerans_Agy99 RCRIQPVFQNPYSSLDPMYSVFRAIEEPLRIHRVGD-RGQRRQMVAELVD
MLBr_01122|M.leprae_Br4923 RRNIQVVFQDPVASLDPRLPVFELIAEPLHANGF-S-KNYTQMRVAELLD
Mflv_2758|M.gilvum_PYR-GCK RRRIGLVFQDPFASLNPRARVDTSIAEPLVVHKVVDGKDARHRRVGELLD
Mvan_3776|M.vanbaalenii_PYR-1 RRRVGLVFQDPFSSLNPRARVGDSVSEPLVVHKLVDGRETRRRRVEDLLD
TH_4070|M.thermoresistible__bu RRRISLVFQDPFSSLNPRMRVIDSISEPLRVHGLADGDRARCRRVADLLE
MAB_0429|M.abscessus_ATCC_1997 RSKVGVVFQDPGSSLNPRWPIGQSIAEPLTLHTDMD-RTQREGRVKTLLE
: *:*:* ::*:* : : ** . : :
MAV_3418|M.avium_104 SVGLSDDKLDRRPGELSGGECQRVAIARALVLRPQIVVLDEPLSALDVVT
MSMEG_2844|M.smegmatis_MC2_155 NVNLSRDKLTRRTVELSGGERQRVAIARALVVQPQIILLDEPLSALDVVT
Mb3687c|M.bovis_AF2122/97 QVALPSSILGRRPRELSGGQRQRVAIARALALRPEVLVCDEAVSALDVLV
Rv3663c|M.tuberculosis_H37Rv QVALPSSILGRRPRELSGGQRQRVAIARALALRPEVLVCDEAVSALDVLV
MMAR_5151|M.marinum_M QVALPSSVLGRLPRELSGGQRQRVAIARALALRPDVLVCDEAVSALDVLV
MUL_4237|M.ulcerans_Agy99 QVALPSSVLGRLPRELSGGQRQRVAIARALALRPDVLVCDEAVSALDVLV
MLBr_01122|M.leprae_Br4923 IVGLRRDDATRYPTEFSGGQKQRIGIARALALQPKILAFDEPVSALDVSI
Mflv_2758|M.gilvum_PYR-GCK LVGLPAEFAQRYPHELSGGQRQRVSIARALAAEPDLLILDEATASLDVSV
Mvan_3776|M.vanbaalenii_PYR-1 LVGLPTEFADRYPHELSGGQRQRVSIARALAAEPDLLILDEATASLDVSA
TH_4070|M.thermoresistible__bu LVGLPADFGARYPHELSGGQRQRVSVARALATDPDLLILDESTASLDVSV
MAB_0429|M.abscessus_ATCC_1997 QVQLPAAMRNRFPHQLSGGQRQRVGIARALALEPTLLIADEPTSALDVSV
* * * . ::***: **:.:****. * :: **. ::***
MAV_3418|M.avium_104 QDQIVGLLLRLQRQLGLTYLFVSHDLSVVRRLAHRVVVLRAGRVVETGDT
MSMEG_2844|M.smegmatis_MC2_155 QDQILGLLRELQDNLGLTYLFISHDLSVVQQLSHRVVVLKAGQVAESGDT
Mb3687c|M.bovis_AF2122/97 QAQILDLLADLQADLGLTYLFISHDLAVIRQIADDVLVMRAGRVVEHAST
Rv3663c|M.tuberculosis_H37Rv QAQILDLLADLQADLGLTYLFISHDLAVIRQIADDVLVMRAGRVVEHAST
MMAR_5151|M.marinum_M QAQILDLLAELQADLGLAYLFISHDLAVVRQISDDVLVMRAGQVVEHAPT
MUL_4237|M.ulcerans_Agy99 QAQILDLLAELQADLGLAYLFISHDLAVVRQISDDVLVMRAGQVVEHAPT
MLBr_01122|M.leprae_Br4923 QAGIINLLLDLQDRLGLSYLFVSHDLSVIKHLAHQVAVMLAGTIVEQGDS
Mflv_2758|M.gilvum_PYR-GCK QARVLELLSRLQSELALTYLLIGHDLAVIRQLSHDVLVMRDGEAVENRPA
Mvan_3776|M.vanbaalenii_PYR-1 QARVLDLLSRLQRELGLTYLLIGHDLAVIRQVSHEILVMRGGQVVENRPA
TH_4070|M.thermoresistible__bu QDRVLDLLADLQRELGLTYLFIAHDLAVVERMSHDVLVLRAGETVEYRPA
MAB_0429|M.abscessus_ATCC_1997 QATVLDLFAELQREHGFACLFISHDLAVVELVASRIAVLNRGRLAEFGSD
* :: *: ** .:: *::.***:*:. :: : *: * .*
MAV_3418|M.avium_104 AAVFAAPAADYTRALLDAVPGRRLAH-------PALSPS
MSMEG_2844|M.smegmatis_MC2_155 AAVFSAPTSPYTRALIDAVPGRRLAR-------LALAS-
Mb3687c|M.bovis_AF2122/97 EEVFSRPRHEYTRQLLQAIPGAPSAP-------RKVGNL
Rv3663c|M.tuberculosis_H37Rv EEVFSRPRHEYTRQLLQAIPGAPSAP-------RKVGNL
MMAR_5151|M.marinum_M EEVFTRPGAQYTQRLLEAIPGGPVAP-------R-----
MUL_4237|M.ulcerans_Agy99 EEVFTRPGAQYTQRLLEAIPGGPVAP-------R-----
MLBr_01122|M.leprae_Br4923 HDVFRNPQHDYTRRLLGAIPRPEVG--------------
Mflv_2758|M.gilvum_PYR-GCK DDLFAHPEQQYTRDLLAAVPPRTP---------RAAV--
Mvan_3776|M.vanbaalenii_PYR-1 DELFTSPQQEYTRALLAAVPPVKP---------RSAV--
TH_4070|M.thermoresistible__bu AELFAAPEHEYTRALLAAVPAVRP---------RAAG--
MAB_0429|M.abscessus_ATCC_1997 TQVLTAPKDDYTKRLLAAVPVPDPEQQRIRREERQALR-
:: * **: *: *:*