For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NMGMVTLAAIRSVPMATTYIRGGTVVSPTGCQDLDVLIDGERIAALVDPELAPFDDPDRTFDHVLEAKGK YVIPGGVDAHTHMEMPFGGTVASDTFETGTRAAAFGGTTTIIDFAIQRQGERVQDTLAAWHAKARGNCAV DYGFHQILGGVDTDALKAMDELVAEGVTSFKLFMAYPGVYYSDDGQILRAMQQCAGNGALTMMHAENGIA IDQLVAQHIERGETSPYYHGVSRPWELEEEATHRAIMLAKITGAPLYVVHVSAKQAMEQIAGARGSGANV FAETCPQYLYLSLEEQLAAPGFEGAKWVCSTPLRSHEEGHQEELWRYIRTGDVATVSTDHCPFCFKGQKE MGLGDFSKIPNGMSGVEHRIDLMYQGVVNGKLSLQRWVDVCATTPARMFGLYPRKGIVSPGADADVVIYD PDGRTSIGFEKNHHSNVDHSTWEGFEVEGCVQTVLSRGTIVVDRGRYLGRKGHGVFLKRGLSQYLI*
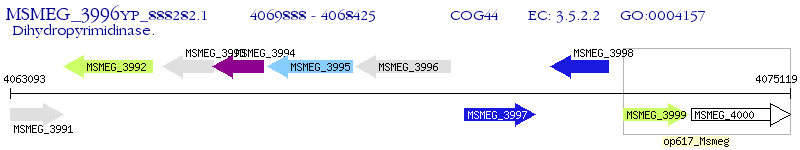
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3996 | hydA | - | 100% (487) | dihydropyrimidinase |
| M. smegmatis MC2 155 | MSMEG_4012 | hydA | 0.0 | 75.90% (473) | dihydropyrimidinase |
| M. smegmatis MC2 155 | MSMEG_3553 | hydA | 0.0 | 67.58% (472) | dihydropyrimidinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1416 | pyrC | 9e-24 | 26.14% (440) | dihydroorotase |
| M. gilvum PYR-GCK | Mflv_3169 | - | 2e-40 | 30.74% (462) | allantoinase |
| M. tuberculosis H37Rv | Rv1381 | pyrC | 7e-24 | 26.36% (440) | dihydroorotase |
| M. leprae Br4923 | MLBr_00533 | pyrC | 4e-25 | 26.98% (441) | dihydroorotase |
| M. abscessus ATCC 19977 | MAB_2817c | - | 0.0 | 67.02% (473) | phenylhydantoinase |
| M. marinum M | MMAR_2196 | pyrC | 3e-24 | 27.07% (447) | dihydroorotase PyrC |
| M. avium 104 | MAV_3392 | pyrC | 5e-26 | 26.83% (436) | dihydroorotase |
| M. thermoresistible (build 8) | TH_3497 | - | 3e-09 | 28.65% (178) | PUTATIVE acyl-CoA thioesterase |
| M. ulcerans Agy99 | MUL_1781 | pyrC | 5e-24 | 26.14% (440) | dihydroorotase |
| M. vanbaalenii PYR-1 | Mvan_2661 | pyrC | 4e-27 | 27.06% (436) | dihydroorotase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00533|M.leprae_Br4923 ----MS------------VLLRG--VRLYGEGEQVDVLVEGEQIANIGAG
MAV_3392|M.avium_104 ----MS------------VLLRG--VRLYGEGDRVDVLVDDGQIADIGAG
Mb1416|M.bovis_AF2122/97 ----MS------------VLIRG--VRPYGEGERVDVLVDDGQIAQIGPD
Rv1381|M.tuberculosis_H37Rv ----MS------------VLIRG--VRPYGEGERVDVLVDDGQIAQIGPD
MMAR_2196|M.marinum_M ----MS------------VLLRG--VRLYGEGERVDVLVDDNQIADIGTG
MUL_1781|M.ulcerans_Agy99 ----MS------------VLLRG--VRLYGEGERVDVLVDDNQIADIGTG
Mvan_2661|M.vanbaalenii_PYR-1 ----MTSSSPESLRSCPPIVLKS--VRLYGEGDPVDVLVRDGQIAEIGER
TH_3497|M.thermoresistible__bu -------------------LIRN--VRPYGEGEPVDVLVADGQIAEIGEK
MSMEG_3996|M.smegmatis_MC2_155 MNMGMVTLAAIRSVPMATTYIRGGTVVSPTGCQDLDVLIDGERIAALVDP
MAB_2817c|M.abscessus_ATCC_199 ---------------MATTLIRGGTVVSATGRGAADVLVDGATIAAVLLP
Mflv_3169|M.gilvum_PYR-GCK ---MTADTREQSNHADLDLVVRGERMLTTAGIVAREIGIRDGRIVAIEPL
::. : :: : . *. :
MLBr_00533|M.leprae_Br4923 IDIP-------DTADVIDAIDQVLLPGLVDLHTHLREP--GREYAEDIET
MAV_3392|M.avium_104 LDIP-------DTADVIDATGQVLLPGFVDLHTHLREP--GREYAEDIET
Mb1416|M.bovis_AF2122/97 LAIP-------DTADVIDATGHVLLPGFVDLHTHLREP--GREYAEDIET
Rv1381|M.tuberculosis_H37Rv LAIP-------DTADVIDATGHVLLPGFVDLHTHLREP--GREYAEDIET
MMAR_2196|M.marinum_M LAIP-------DTADVIDATDHVLLPGLVDLHTHLREP--GREYAEDIET
MUL_1781|M.ulcerans_Agy99 LAIP-------DTADVIDATDHVLLPGLVDLHTHLREP--GREYAEDIET
Mvan_2661|M.vanbaalenii_PYR-1 LTAP-------TDADSIDCEGKILLPGFVDLHTHLREP--GREYAEDIET
TH_3497|M.thermoresistible__bu LPIPGD----GELGDVIDADGQILLPGFVDLHTHLREP--GREYAEDIET
MSMEG_3996|M.smegmatis_MC2_155 ELAPFD-DPDRTFDHVLEAKGKYVIPGGVDAHTHMEMPFGGTVASDTFET
MAB_2817c|M.abscessus_ATCC_199 GSTLLGMDLAKTVDTVIEAHGKYVIPGGIDAHTHMSMPFGGTTASDDFET
Mflv_3169|M.gilvum_PYR-GCK GSGLPG------AEIVELTDEQVMIPGLVDTHVHVNEP--GRTEWEGFDS
: ::** :* *.*: * * : :::
MLBr_00533|M.leprae_Br4923 GSAAAALGGYTAVFAMPNTTPVADSPVVTDHVWRRGQQVGLVDVHPVGAV
MAV_3392|M.avium_104 GSAAAALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAV
Mb1416|M.bovis_AF2122/97 GSAAAALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAV
Rv1381|M.tuberculosis_H37Rv GSAAAALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAV
MMAR_2196|M.marinum_M GSAAAALGGYTAVFAMANTHPVADSPVVTDHVWHRGQQVGLVDVHPVGAV
MUL_1781|M.ulcerans_Agy99 GSAAAALGGYTAVFAMANTHPVADSPVVTDHVWHRGQQVGLVDVHPVGAV
Mvan_2661|M.vanbaalenii_PYR-1 GSAAAALGGYTAVFAMANTDPVADSAVVTDHVWHRGQQVGLVDVHPVGAV
TH_3497|M.thermoresistible__bu GSAAAALGGYTAVFAMANTDPVADSPVVTDHVWRRGQQVGLVDVHPVGAV
MSMEG_3996|M.smegmatis_MC2_155 GTRAAAFGGTTTIIDFAIQR-QGERVQDTLAAWHAKARGNCAVDYGFHQI
MAB_2817c|M.abscessus_ATCC_199 GTRAAAWGGTTTIIDFAVQK-FGERLQDSLDTWHEMAAGQCAIDYSFHQI
Mflv_3169|M.gilvum_PYR-GCK ATRAAAAGGVTTLIDMPLNSIPPTVNVDALNAKREAASGKLHIDVGFWGG
.: *** ** *::: :. : . : .
MLBr_00533|M.leprae_Br4923 TVGLAGAELTEMG-MMAAGAAQVRIFSDD-GNCVHNPLVMRRALEYATGL
MAV_3392|M.avium_104 TMGLAGKELTEMG-MMAAGAAQVRMFSDD-GVCVHDPLVMRRALEYATGL
Mb1416|M.bovis_AF2122/97 TVGLAGAELTEMG-MMNAGAAQVRMFSDD-GVCVHDPLIMRRALEYATGL
Rv1381|M.tuberculosis_H37Rv TVGLAGAELTEMG-MMNAGAAQVRMFSDD-GVCVHDPLIMRRALEYATGL
MMAR_2196|M.marinum_M TVGLAGTELTEMG-MMNAGVAAVRMFSDD-GICVHDPLIMRRALEYATGL
MUL_1781|M.ulcerans_Agy99 TIGLAGTELTEMG-MMNAGVAAVRMFSDD-GICVHDPLIMRRALEYATGL
Mvan_2661|M.vanbaalenii_PYR-1 TVGLDGKQLTEMG-LMAAGAGQVRMFSDD-GVCVDDPLVMRRALEYASGL
TH_3497|M.thermoresistible__bu TVGLEGRQLTEMG-LMAAGAARVRMFSDD-GLCVHDPLVMRRALEYARGL
MSMEG_3996|M.smegmatis_MC2_155 LGGVDTDALKAMDELVAEGVTSFKLFMAYPGVYYSDDGQILRAMQQCAGN
MAB_2817c|M.abscessus_ATCC_199 VGDVNDASLAALRRLTDEGITSYKMFMAYPGVFYSDDAQIVRAMQVGAAT
Mflv_3169|M.gilvum_PYR-GCK AIPGNTGDLRGLHDDGVFGFKCFLLHSGVDEFPHLDADEMEEDMRVLVGF
* : * :. : : . :. .
MLBr_00533|M.leprae_Br4923 GVLIAQHAEEPRLTVGAVAHE-----------------------------
MAV_3392|M.avium_104 GVLIAQHAEEPRLTVGAVAHE-----------------------------
Mb1416|M.bovis_AF2122/97 GVLIAQHAEEPRLTVGAFAHE-----------------------------
Rv1381|M.tuberculosis_H37Rv GVLIAQHAEEPRLTVGAVAHE-----------------------------
MMAR_2196|M.marinum_M GVLIAQHAEEPRLTVGAVAHE-----------------------------
MUL_1781|M.ulcerans_Agy99 GVLIAQHAEEPRLTVGAVAHE-----------------------------
Mvan_2661|M.vanbaalenii_PYR-1 GVLIAQHAEEPRLTVGAVAHE-----------------------------
TH_3497|M.thermoresistible__bu GVLIAQHAEEPRLTVGAVAHXXXWPACPARCRPGYRSPPTWCASAPPGRP
MSMEG_3996|M.smegmatis_MC2_155 GALTMMHAENGIAIDQLVAQHIE---------------------------
MAB_2817c|M.abscessus_ATCC_199 GLMTMMHAENGPAIDVLVAQLLA---------------------------
Mflv_3169|M.gilvum_PYR-GCK DSMMIVHAEDSRAIDHAPTAE-----------------------------
. : ***: :
MLBr_00533|M.leprae_Br4923 -------------GPT-----------AARLGLSGWPR------------
MAV_3392|M.avium_104 -------------GPT-----------AARLGLAGWPR------------
Mb1416|M.bovis_AF2122/97 -------------GPM-----------AARLGLAGWPR------------
Rv1381|M.tuberculosis_H37Rv -------------GPM-----------AARLGLAGWPR------------
MMAR_2196|M.marinum_M -------------GPN-----------AAQLGLAGWPR------------
MUL_1781|M.ulcerans_Agy99 -------------GPS-----------AAQLGLAGWPR------------
Mvan_2661|M.vanbaalenii_PYR-1 -------------GPN-----------AARLGLAGWPR------------
TH_3497|M.thermoresistible__bu NAVRRNRSSPRRIGPTPPPGTSRPATDVGRLGSADRARRPGPRCRGAWWL
MSMEG_3996|M.smegmatis_MC2_155 ------------RGET-------------SPYYHGVSR------------
MAB_2817c|M.abscessus_ATCC_199 ------------EGKT-------------DPYYHGIAR------------
Mflv_3169|M.gilvum_PYR-GCK -----------GDR--------------YSRFLASRPR------------
. .*
MLBr_00533|M.leprae_Br4923 ---AAEESIVARDALLARDAG--------------ARVHICHASTAGTVE
MAV_3392|M.avium_104 ---AAEESIVARDALLARDAG--------------ARVHICHASTAGTVE
Mb1416|M.bovis_AF2122/97 ---AAEESIVARDALLARDAG--------------ARVHICHASAAGTVE
Rv1381|M.tuberculosis_H37Rv ---AAEESIVARDALLARDAG--------------ARVHICHASAAGTVE
MMAR_2196|M.marinum_M ---AAEESIVARDALLARDAG--------------ARVHICHASTVGTVE
MUL_1781|M.ulcerans_Agy99 ---AAEESIVARDALLTRDAG--------------ARVHICHASTVGTVE
Mvan_2661|M.vanbaalenii_PYR-1 ---AAEESIVARDALLARDAG--------------ARVHICHASTAGTVE
TH_3497|M.thermoresistible__bu GHDAPVTELTLYDVLTTLDVAQVDDDLFEATQLDNPAHHIVGGHIAG-QA
MSMEG_3996|M.smegmatis_MC2_155 -PWELEEEATHRAIMLAKITG--------------APLYVVHVSAKQAME
MAB_2817c|M.abscessus_ATCC_199 -AWQLEEEATHRAIMLSNLTG--------------APLYVVHVSAKQAVA
Mflv_3169|M.gilvum_PYR-GCK ---GAENVAIAEVIERARWTG--------------ARAHILHLSSSDALP
: .. . ::
MLBr_00533|M.leprae_Br4923 ILKWAKEQGISITAEVTPHHLLLDDSRLASYDGVNRVN-------PPLRE
MAV_3392|M.avium_104 IVKWAKQQGISITAEVTPHHLMLDDSRLASYDGRNRVN-------PPLRE
Mb1416|M.bovis_AF2122/97 ILKWAKDQGISITAEVTPHHLLLDDARLASYDGVNRVN-------PPLRE
Rv1381|M.tuberculosis_H37Rv ILKWAKDQGISITAEVTPHHLLLDDARLASYDGVNRVN-------PPLRE
MMAR_2196|M.marinum_M ILKWAKSQGISITAEVTPHHLLLDDRRLSGYDGVNRVN-------PPLRE
MUL_1781|M.ulcerans_Agy99 ILKWAKSQGISITAEVTPHHLLLDDRRLSGYDGVNRVN-------PPLRE
Mvan_2661|M.vanbaalenii_PYR-1 LLKWAKSQGISITAEVTPHHLLLDDSRLDTYDGVNRVN-------PPLRE
TH_3497|M.thermoresistible__bu VVAASRTAPQRTAHSAHVYFLRAGDARYPVQMRVDRLRDGGSLSTXRVTA
MSMEG_3996|M.smegmatis_MC2_155 QIAGARGSGANVFAETCPQYLYLSLEEQLAAPGFEGAKWVCSTP-LRSHE
MAB_2817c|M.abscessus_ATCC_199 QLAAARDAGQNVYGETCPQYLYLSLEDNLGAPGFDGAKWVCSTP-LRAKH
Mflv_3169|M.gilvum_PYR-GCK MIATAKRDGVRITVETCPHYLTLLAEEIPNGATAFKCC-------PPIRE
: :: .. .*
MLBr_00533|M.leprae_Br4923 TADAVALRQALADG--IIDCVTTDHAP---------HADHEKCVEFSAAR
MAV_3392|M.avium_104 AADAAALRRALADG--VIDCVATDHAP---------HAEHEKCVEFSAAR
Mb1416|M.bovis_AF2122/97 ASDAVALRQALADG--IIDCVATDHAP---------HAEHEKCVEFAAAR
Rv1381|M.tuberculosis_H37Rv ASDAVALRQALADG--IIDCVATDHAP---------HAEHEKCVEFAAAR
MMAR_2196|M.marinum_M ASDAMALRQALADG--IIDCVATDHAP---------HAEHEKCVEFSAAR
MUL_1781|M.ulcerans_Agy99 ASDAMALRQALADG--IIDCVATDHAP---------HAEHEKCVEFSAAR
Mvan_2661|M.vanbaalenii_PYR-1 VSDAVALRRALADG--VIDCVATDHAP---------HGAHEKCCEFANAR
TH_3497|M.thermoresistible__bu TQDGQVLLEALASFTVRIDSVDTHHPMPDVPDPDTLPPVTEQLVAYADEF
MSMEG_3996|M.smegmatis_MC2_155 EGHQEELWRYIRTG--DVATVSTDHCPFCFK-----GQKEMGLGDFSKIP
MAB_2817c|M.abscessus_ATCC_199 EHHQDEMWRALRTN--DIQMVSTDHCPFCMK-----GQKDMGIGDFSKIP
Mflv_3169|M.gilvum_PYR-GCK ASNRELLWQGLIDG--TIDCIVSDHSPS------TIDLKDVENGDFGVAW
. : . : : : :.* :.
MLBr_00533|M.leprae_Br4923 PGMLGLQTALS--------VVVHTMVVPGLLS--WRDVAQVMSENPARIA
MAV_3392|M.avium_104 PGMLGLQTALS--------IVVHTMVRPGLLT--WRDVAKVMSENPARIA
Mb1416|M.bovis_AF2122/97 PGMLGLQTALS--------VVVQTMVAPGLLS--WRDIARVMSENPACIA
Rv1381|M.tuberculosis_H37Rv PGMLGLQTALS--------VVVQTMVAPGLLS--WRDIARVMSENPACIA
MMAR_2196|M.marinum_M PGMLGLQTALS--------VVVQTMVQPGLLS--WRDVARVMSENPARIV
MUL_1781|M.ulcerans_Agy99 PGMLGLQTALS--------VVVQTMVQPGLLS--WRDVARVMSENPARIV
Mvan_2661|M.vanbaalenii_PYR-1 PGMLGLQTALS--------VVVETLVNPGLLT--WRDVARVMSEAPARIV
TH_3497|M.thermoresistible__bu GGLWVRPQPIERRYIDPPPRVAYDLPEPPARTRMWWRATEPVPDDPALNS
MSMEG_3996|M.smegmatis_MC2_155 NGMSGVEHRID---------LMYQGVVNGKLS--LQRWVDVCATTPARMF
MAB_2817c|M.abscessus_ATCC_199 NGIGSIEHRMD---------LLYQGVVDGRIS--VERWVEITSTTPARMF
Mflv_3169|M.gilvum_PYR-GCK GGVASLQLGLS---------LIWTEAKRRGVA--LTRVIDWMAAKPAELA
*: :. : : . **
MLBr_00533|M.leprae_Br4923 GLPDHGR---PLEVGEPAN-------LTVVDPDVTWTVTGDGLASRSANT
MAV_3392|M.avium_104 NLPDHGR---PLEVGEPAN-------LTVVDPDASWTVAGSDLASRSANT
Mb1416|M.bovis_AF2122/97 RLPDQGR---PLEVGEPAN-------LTVVDPDATWTVTGADLASRSANT
Rv1381|M.tuberculosis_H37Rv RLPDQGR---PLEVGEPAN-------LTVVDPDATWTVTGADLASRSANT
MMAR_2196|M.marinum_M GLPDHGR---PLEVGEPAN-------LTVIDPDATWTVTGADLASRSANT
MUL_1781|M.ulcerans_Agy99 GLPDHGR---PLEVGEPAN-------LTVIDPDATWTVTGADLASRSANT
Mvan_2661|M.vanbaalenii_PYR-1 GLPDQGR---PLEVGEPAN-------LTVVDPAARWTVKGPELASRSDNT
TH_3497|M.thermoresistible__bu ALLTYVTGTSTLEAAMVAHRTTPLHSFNALIDHALWFHRPAHLTDWVLAD
MSMEG_3996|M.smegmatis_MC2_155 GLYPRKGIVSPGADADVVIYDPDGRTSIGFEKNHHSNVDHSTWEGFEVEG
MAB_2817c|M.abscessus_ATCC_199 GLYGRKGVIAPGADADIVVYDPNGHTSIGLGKTHHMNMDHSAWEGYEIDG
Mflv_3169|M.gilvum_PYR-GCK GLNNKGK-IALGYDADFAIFEPESAQVVDVHKLHHRNPITP-YDGRAVAG
* . . . .
MLBr_00533|M.leprae_Br4923 PYEAMTLPAT--VTATLLRGK--------------VTARGQKSPV--
MAV_3392|M.avium_104 PYEAMTLPAT--VTATLLRGK--------------VTARDGKCPV--
Mb1416|M.bovis_AF2122/97 PFESMSLPAT--VTATLLRGK--------------VTARDGKIRA--
Rv1381|M.tuberculosis_H37Rv PFESMSLPAT--VTATLLRGK--------------VTARDGKIRA--
MMAR_2196|M.marinum_M PYESMTLPAR--VTATLLRGK--------------ITARDGKSPA--
MUL_1781|M.ulcerans_Agy99 PYESMTLPAR--VTATLLRGK--------------ITARDGKSPA--
Mvan_2661|M.vanbaalenii_PYR-1 PYEDMELPAV--VTATLLRGK--------------VTARDGKVGA--
TH_3497|M.thermoresistible__bu QFSPSSLGGRGLATATMYNRTGELVCTATQELYFGRPARSGSRR---
MSMEG_3996|M.smegmatis_MC2_155 CVQTVLSRGTIVVDRGRYLGRKG----------HGVFLKRGLSQYLI
MAB_2817c|M.abscessus_ATCC_199 HVDTVLSRGKVIVDGDEYLGSKG----------DGVFLRRDLCQYLI
Mflv_3169|M.gilvum_PYR-GCK VVASTWLRGT-KIDFTTPRGR---------------MLRRGGV----
. :