For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSTLITGGTIVNATGTCTADVLVDGERIAAVFAPGTSLGNHPPVVDRTIDATGKYVIPGGVDAHTHMEM PFGGTNASDTFETGTRAAAWGGTTTIVDFVVQKAGENPLVQYERWQEKAAGKCAIDYGFHQILSDVDDSS LSAMDELVREGVTSFKFFMAYKGVFLSDDGQILRGMQKARDNGALIMMHAENGSVIDVLVQQSVARGDTS PYHHGLTRPWQAEAEATHRAIMLADITGAPLYIVHVSAKQAVEQIAMARDNGRNVFAETCPQYLYLSLEE HLGAPGFEGAKWVCSTPLRSRAEHHQQHMWQGLRTNDLQVVSTDHCPFCMRGQKELGRDDFSKIPNGIGS VEHRMDLLYQGVVLGEISLPRWVEICSTTPARMFGMYGRKGVIQPGADADIVIYDPKGKTRISAETHHMN IDHSAWEGFEVDGHVDTVLSRGRVIVDENSFHGQAGHGEYLKRSLSQYLI
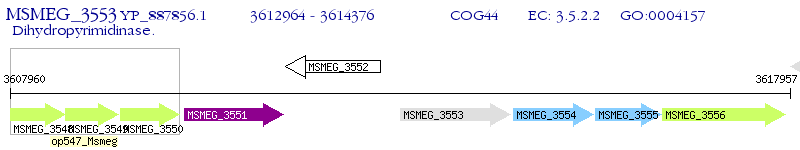
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3553 | hydA | - | 100% (470) | dihydropyrimidinase |
| M. smegmatis MC2 155 | MSMEG_3996 | hydA | 0.0 | 67.58% (472) | dihydropyrimidinase |
| M. smegmatis MC2 155 | MSMEG_4012 | hydA | 0.0 | 67.86% (473) | dihydropyrimidinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1416 | pyrC | 1e-21 | 25.29% (427) | dihydroorotase |
| M. gilvum PYR-GCK | Mflv_3169 | - | 4e-35 | 28.04% (453) | allantoinase |
| M. tuberculosis H37Rv | Rv1381 | pyrC | 3e-22 | 25.53% (427) | dihydroorotase |
| M. leprae Br4923 | MLBr_00533 | pyrC | 3e-23 | 25.92% (436) | dihydroorotase |
| M. abscessus ATCC 19977 | MAB_2817c | - | 0.0 | 69.13% (473) | phenylhydantoinase |
| M. marinum M | MMAR_2196 | pyrC | 3e-24 | 26.00% (427) | dihydroorotase PyrC |
| M. avium 104 | MAV_3392 | pyrC | 2e-22 | 25.29% (427) | dihydroorotase |
| M. thermoresistible (build 8) | TH_3497 | - | 4e-09 | 26.86% (175) | PUTATIVE acyl-CoA thioesterase |
| M. ulcerans Agy99 | MUL_1781 | pyrC | 3e-24 | 26.00% (427) | dihydroorotase |
| M. vanbaalenii PYR-1 | Mvan_2661 | pyrC | 1e-21 | 25.53% (427) | dihydroorotase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00533|M.leprae_Br4923 -MS------------VLLRG--VRLYGEGEQVDVLVEGEQIANIGAGIDI
MAV_3392|M.avium_104 -MS------------VLLRG--VRLYGEGDRVDVLVDDGQIADIGAGLDI
Mb1416|M.bovis_AF2122/97 -MS------------VLIRG--VRPYGEGERVDVLVDDGQIAQIGPDLAI
Rv1381|M.tuberculosis_H37Rv -MS------------VLIRG--VRPYGEGERVDVLVDDGQIAQIGPDLAI
MMAR_2196|M.marinum_M -MS------------VLLRG--VRLYGEGERVDVLVDDNQIADIGTGLAI
MUL_1781|M.ulcerans_Agy99 -MS------------VLLRG--VRLYGEGERVDVLVDDNQIADIGTGLAI
Mvan_2661|M.vanbaalenii_PYR-1 -MTSSSPESLRSCPPIVLKS--VRLYGEGDPVDVLVRDGQIAEIGERLTA
TH_3497|M.thermoresistible__bu ----------------LIRN--VRPYGEGEPVDVLVADGQIAEIGEKLPI
MSMEG_3553|M.smegmatis_MC2_155 ------------MSSTLITGGTIVNATGTCTADVLVDGERIAAVFAPGTS
MAB_2817c|M.abscessus_ATCC_199 ------------MATTLIRGGTVVSATGRGAADVLVDGATIAAVLLPGST
Mflv_3169|M.gilvum_PYR-GCK MTADTREQSNHADLDLVVRGERMLTTAGIVAREIGIRDGRIVAIEPLGSG
:: . : :: : . *. :
MLBr_00533|M.leprae_Br4923 P-------DTADVIDAIDQVLLPGLVDLHTHLREP--GREYAEDIETGSA
MAV_3392|M.avium_104 P-------DTADVIDATGQVLLPGFVDLHTHLREP--GREYAEDIETGSA
Mb1416|M.bovis_AF2122/97 P-------DTADVIDATGHVLLPGFVDLHTHLREP--GREYAEDIETGSA
Rv1381|M.tuberculosis_H37Rv P-------DTADVIDATGHVLLPGFVDLHTHLREP--GREYAEDIETGSA
MMAR_2196|M.marinum_M P-------DTADVIDATDHVLLPGLVDLHTHLREP--GREYAEDIETGSA
MUL_1781|M.ulcerans_Agy99 P-------DTADVIDATDHVLLPGLVDLHTHLREP--GREYAEDIETGSA
Mvan_2661|M.vanbaalenii_PYR-1 P-------TDADSIDCEGKILLPGFVDLHTHLREP--GREYAEDIETGSA
TH_3497|M.thermoresistible__bu PG----DGELGDVIDADGQILLPGFVDLHTHLREP--GREYAEDIETGSA
MSMEG_3553|M.smegmatis_MC2_155 LG--NHPPVVDRTIDATGKYVIPGGVDAHTHMEMPFGGTNASDTFETGTR
MAB_2817c|M.abscessus_ATCC_199 LLGMDLAKTVDTVIEAHGKYVIPGGIDAHTHMSMPFGGTTASDDFETGTR
Mflv_3169|M.gilvum_PYR-GCK LP------GAEIVELTDEQVMIPGLVDTHVHVNEP--GRTEWEGFDSATR
: ::** :* *.*: * * : :::.:
MLBr_00533|M.leprae_Br4923 AAALGGYTAVFAMPNTTPVADSPVVTDHVWRRGQQVGLVDVHPVGAVTVG
MAV_3392|M.avium_104 AAALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTMG
Mb1416|M.bovis_AF2122/97 AAALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTVG
Rv1381|M.tuberculosis_H37Rv AAALGGYTAVFAMANTNPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTVG
MMAR_2196|M.marinum_M AAALGGYTAVFAMANTHPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTVG
MUL_1781|M.ulcerans_Agy99 AAALGGYTAVFAMANTHPVADSPVVTDHVWHRGQQVGLVDVHPVGAVTIG
Mvan_2661|M.vanbaalenii_PYR-1 AAALGGYTAVFAMANTDPVADSAVVTDHVWHRGQQVGLVDVHPVGAVTVG
TH_3497|M.thermoresistible__bu AAALGGYTAVFAMANTDPVADSPVVTDHVWRRGQQVGLVDVHPVGAVTVG
MSMEG_3553|M.smegmatis_MC2_155 AAAWGGTTTIVDFV-VQKAGENPLVQYERWQEKAAGKCAIDYGFHQILSD
MAB_2817c|M.abscessus_ATCC_199 AAAWGGTTTIIDFA-VQKFGERLQDSLDTWHEMAAGQCAIDYSFHQIVGD
Mflv_3169|M.gilvum_PYR-GCK AAAAGGVTTLIDMPLNSIPPTVNVDALNAKREAASGKLHIDVGFWGGAIP
*** ** *::. : . :. .
MLBr_00533|M.leprae_Br4923 LAGAELTEMG-MMAAGAAQVRIFSDD-GNCVHNPLVMRRALEYATGLGVL
MAV_3392|M.avium_104 LAGKELTEMG-MMAAGAAQVRMFSDD-GVCVHDPLVMRRALEYATGLGVL
Mb1416|M.bovis_AF2122/97 LAGAELTEMG-MMNAGAAQVRMFSDD-GVCVHDPLIMRRALEYATGLGVL
Rv1381|M.tuberculosis_H37Rv LAGAELTEMG-MMNAGAAQVRMFSDD-GVCVHDPLIMRRALEYATGLGVL
MMAR_2196|M.marinum_M LAGTELTEMG-MMNAGVAAVRMFSDD-GICVHDPLIMRRALEYATGLGVL
MUL_1781|M.ulcerans_Agy99 LAGTELTEMG-MMNAGVAAVRMFSDD-GICVHDPLIMRRALEYATGLGVL
Mvan_2661|M.vanbaalenii_PYR-1 LDGKQLTEMG-LMAAGAGQVRMFSDD-GVCVDDPLVMRRALEYASGLGVL
TH_3497|M.thermoresistible__bu LEGRQLTEMG-LMAAGAARVRMFSDD-GLCVHDPLVMRRALEYARGLGVL
MSMEG_3553|M.smegmatis_MC2_155 VDDSSLSAMDELVREGVTSFKFFMAYKGVFLSDDGQILRGMQKARDNGAL
MAB_2817c|M.abscessus_ATCC_199 VNDASLAALRRLTDEGITSYKMFMAYPGVFYSDDAQIVRAMQVGAATGLM
Mflv_3169|M.gilvum_PYR-GCK GNTGDLRGLHDDGVFGFKCFLLHSGVDEFPHLDADEMEEDMRVLVGFDSM
.* : * :. : : . :. . :
MLBr_00533|M.leprae_Br4923 IAQHAEEPRLTVGAVAHE--------------------------------
MAV_3392|M.avium_104 IAQHAEEPRLTVGAVAHE--------------------------------
Mb1416|M.bovis_AF2122/97 IAQHAEEPRLTVGAFAHE--------------------------------
Rv1381|M.tuberculosis_H37Rv IAQHAEEPRLTVGAVAHE--------------------------------
MMAR_2196|M.marinum_M IAQHAEEPRLTVGAVAHE--------------------------------
MUL_1781|M.ulcerans_Agy99 IAQHAEEPRLTVGAVAHE--------------------------------
Mvan_2661|M.vanbaalenii_PYR-1 IAQHAEEPRLTVGAVAHE--------------------------------
TH_3497|M.thermoresistible__bu IAQHAEEPRLTVGAVAHXXXWPACPARCRPGYRSPPTWCASAPPGRPNAV
MSMEG_3553|M.smegmatis_MC2_155 IMMHAENGSVIDVLVQQS--------------------------------
MAB_2817c|M.abscessus_ATCC_199 TMMHAENGPAIDVLVAQL--------------------------------
Mflv_3169|M.gilvum_PYR-GCK MIVHAEDSRAIDHAPTAE--------------------------------
***:
MLBr_00533|M.leprae_Br4923 ----------GPT-----------AARLGLSGWPR---------------
MAV_3392|M.avium_104 ----------GPT-----------AARLGLAGWPR---------------
Mb1416|M.bovis_AF2122/97 ----------GPM-----------AARLGLAGWPR---------------
Rv1381|M.tuberculosis_H37Rv ----------GPM-----------AARLGLAGWPR---------------
MMAR_2196|M.marinum_M ----------GPN-----------AAQLGLAGWPR---------------
MUL_1781|M.ulcerans_Agy99 ----------GPS-----------AAQLGLAGWPR---------------
Mvan_2661|M.vanbaalenii_PYR-1 ----------GPN-----------AARLGLAGWPR---------------
TH_3497|M.thermoresistible__bu RRNRSSPRRIGPTPPPGTSRPATDVGRLGSADRARRPGPRCRGAWWLGHD
MSMEG_3553|M.smegmatis_MC2_155 ---------VARG-----------DTSPYHHGLTR-------------PW
MAB_2817c|M.abscessus_ATCC_199 ---------LAEG-----------KTDPYYHGIAR-------------AW
Mflv_3169|M.gilvum_PYR-GCK ----------GDR------------YSRFLASRPR---------------
. . .*
MLBr_00533|M.leprae_Br4923 AAEESIVARDALLARDAG--------------ARVHICHASTAGTVEILK
MAV_3392|M.avium_104 AAEESIVARDALLARDAG--------------ARVHICHASTAGTVEIVK
Mb1416|M.bovis_AF2122/97 AAEESIVARDALLARDAG--------------ARVHICHASAAGTVEILK
Rv1381|M.tuberculosis_H37Rv AAEESIVARDALLARDAG--------------ARVHICHASAAGTVEILK
MMAR_2196|M.marinum_M AAEESIVARDALLARDAG--------------ARVHICHASTVGTVEILK
MUL_1781|M.ulcerans_Agy99 AAEESIVARDALLTRDAG--------------ARVHICHASTVGTVEILK
Mvan_2661|M.vanbaalenii_PYR-1 AAEESIVARDALLARDAG--------------ARVHICHASTAGTVELLK
TH_3497|M.thermoresistible__bu APVTELTLYDVLTTLDVAQVDDDLFEATQLDNPAHHIVGGHIAG-QAVVA
MSMEG_3553|M.smegmatis_MC2_155 QAEAEATHRAIMLADITG--------------APLYIVHVSAKQAVEQIA
MAB_2817c|M.abscessus_ATCC_199 QLEEEATHRAIMLSNLTG--------------APLYVVHVSAKQAVAQLA
Mflv_3169|M.gilvum_PYR-GCK GAENVAIAEVIERARWTG--------------ARAHILHLSSSDALPMIA
: .. . :: :
MLBr_00533|M.leprae_Br4923 WAKEQGISITAEVTPHHLLLDDSRLASYDGVNRVN-------PPLRETAD
MAV_3392|M.avium_104 WAKQQGISITAEVTPHHLMLDDSRLASYDGRNRVN-------PPLREAAD
Mb1416|M.bovis_AF2122/97 WAKDQGISITAEVTPHHLLLDDARLASYDGVNRVN-------PPLREASD
Rv1381|M.tuberculosis_H37Rv WAKDQGISITAEVTPHHLLLDDARLASYDGVNRVN-------PPLREASD
MMAR_2196|M.marinum_M WAKSQGISITAEVTPHHLLLDDRRLSGYDGVNRVN-------PPLREASD
MUL_1781|M.ulcerans_Agy99 WAKSQGISITAEVTPHHLLLDDRRLSGYDGVNRVN-------PPLREASD
Mvan_2661|M.vanbaalenii_PYR-1 WAKSQGISITAEVTPHHLLLDDSRLDTYDGVNRVN-------PPLREVSD
TH_3497|M.thermoresistible__bu ASRTAPQRTAHSAHVYFLRAGDARYPVQMRVDRLRDGGSLSTXRVTATQD
MSMEG_3553|M.smegmatis_MC2_155 MARDNGRNVFAETCPQYLYLSLEEHLGAPGFEGAKWVCSTP-LRSRAEHH
MAB_2817c|M.abscessus_ATCC_199 AARDAGQNVYGETCPQYLYLSLEDNLGAPGFDGAKWVCSTP-LRAKHEHH
Mflv_3169|M.gilvum_PYR-GCK TAKRDGVRITVETCPHYLTLLAEEIPNGATAFKCC-------PPIREASN
:: .. .* .
MLBr_00533|M.leprae_Br4923 AVALRQALADG--IIDCVTTDHAP---------HADHEKCVEFSAARPGM
MAV_3392|M.avium_104 AAALRRALADG--VIDCVATDHAP---------HAEHEKCVEFSAARPGM
Mb1416|M.bovis_AF2122/97 AVALRQALADG--IIDCVATDHAP---------HAEHEKCVEFAAARPGM
Rv1381|M.tuberculosis_H37Rv AVALRQALADG--IIDCVATDHAP---------HAEHEKCVEFAAARPGM
MMAR_2196|M.marinum_M AMALRQALADG--IIDCVATDHAP---------HAEHEKCVEFSAARPGM
MUL_1781|M.ulcerans_Agy99 AMALRQALADG--IIDCVATDHAP---------HAEHEKCVEFSAARPGM
Mvan_2661|M.vanbaalenii_PYR-1 AVALRRALADG--VIDCVATDHAP---------HGAHEKCCEFANARPGM
TH_3497|M.thermoresistible__bu GQVLLEALASFTVRIDSVDTHHPMPDVPDPDTLPPVTEQLVAYADEFGGL
MSMEG_3553|M.smegmatis_MC2_155 QQHMWQGLRTN--DLQVVSTDHCPFCMR-----GQKELGRDDFSKIPNGI
MAB_2817c|M.abscessus_ATCC_199 QDEMWRALRTN--DIQMVSTDHCPFCMK-----GQKDMGIGDFSKIPNGI
Mflv_3169|M.gilvum_PYR-GCK RELLWQGLIDG--TIDCIVSDHSPS------TIDLKDVENGDFGVAWGGV
: ..* :: : :.* :. *:
MLBr_00533|M.leprae_Br4923 LGLQTALS--------VVVHTMVVPGLLS--WRDVAQVMSENPARIAGLP
MAV_3392|M.avium_104 LGLQTALS--------IVVHTMVRPGLLT--WRDVAKVMSENPARIANLP
Mb1416|M.bovis_AF2122/97 LGLQTALS--------VVVQTMVAPGLLS--WRDIARVMSENPACIARLP
Rv1381|M.tuberculosis_H37Rv LGLQTALS--------VVVQTMVAPGLLS--WRDIARVMSENPACIARLP
MMAR_2196|M.marinum_M LGLQTALS--------VVVQTMVQPGLLS--WRDVARVMSENPARIVGLP
MUL_1781|M.ulcerans_Agy99 LGLQTALS--------VVVQTMVQPGLLS--WRDVARVMSENPARIVGLP
Mvan_2661|M.vanbaalenii_PYR-1 LGLQTALS--------VVVETLVNPGLLT--WRDVARVMSEAPARIVGLP
TH_3497|M.thermoresistible__bu WVRPQPIERRYIDPPPRVAYDLPEPPARTRMWWRATEPVPDDPALNSALL
MSMEG_3553|M.smegmatis_MC2_155 GSVEHRMD---------LLYQGVVLGEIS--LPRWVEICSTTPARMFGMY
MAB_2817c|M.abscessus_ATCC_199 GSIEHRMD---------LLYQGVVDGRIS--VERWVEITSTTPARMFGLY
Mflv_3169|M.gilvum_PYR-GCK ASLQLGLS---------LIWTEAKRRGVA--LTRVIDWMAAKPAELAGLN
:. : : . ** :
MLBr_00533|M.leprae_Br4923 DHGR---PLEVGEPAN-------LTVVDPDVTWTVTGDGLASRSANTPYE
MAV_3392|M.avium_104 DHGR---PLEVGEPAN-------LTVVDPDASWTVAGSDLASRSANTPYE
Mb1416|M.bovis_AF2122/97 DQGR---PLEVGEPAN-------LTVVDPDATWTVTGADLASRSANTPFE
Rv1381|M.tuberculosis_H37Rv DQGR---PLEVGEPAN-------LTVVDPDATWTVTGADLASRSANTPFE
MMAR_2196|M.marinum_M DHGR---PLEVGEPAN-------LTVIDPDATWTVTGADLASRSANTPYE
MUL_1781|M.ulcerans_Agy99 DHGR---PLEVGEPAN-------LTVIDPDATWTVTGADLASRSANTPYE
Mvan_2661|M.vanbaalenii_PYR-1 DQGR---PLEVGEPAN-------LTVVDPAARWTVKGPELASRSDNTPYE
TH_3497|M.thermoresistible__bu TYVTGTSTLEAAMVAHRTTPLHSFNALIDHALWFHRPAHLTDWVLADQFS
MSMEG_3553|M.smegmatis_MC2_155 GRKGVIQPGADADIVIYDPKGKTRISA-ETHHMNIDHSAWEGFEVDGHVD
MAB_2817c|M.abscessus_ATCC_199 GRKGVIAPGADADIVVYDPNGHTSIGLGKTHHMNMDHSAWEGYEIDGHVD
Mflv_3169|M.gilvum_PYR-GCK NKGK-IALGYDADFAIFEPESAQVVDVHKLHHRNPITP-YDGRAVAGVVA
. . .
MLBr_00533|M.leprae_Br4923 AMTLPAT--VTATLLRGK--------------VTARGQKSPV--
MAV_3392|M.avium_104 AMTLPAT--VTATLLRGK--------------VTARDGKCPV--
Mb1416|M.bovis_AF2122/97 SMSLPAT--VTATLLRGK--------------VTARDGKIRA--
Rv1381|M.tuberculosis_H37Rv SMSLPAT--VTATLLRGK--------------VTARDGKIRA--
MMAR_2196|M.marinum_M SMTLPAR--VTATLLRGK--------------ITARDGKSPA--
MUL_1781|M.ulcerans_Agy99 SMTLPAR--VTATLLRGK--------------ITARDGKSPA--
Mvan_2661|M.vanbaalenii_PYR-1 DMELPAV--VTATLLRGK--------------VTARDGKVGA--
TH_3497|M.thermoresistible__bu PSSLGGRGLATATMYNRTGELVCTATQELYFGRPARSGSRR---
MSMEG_3553|M.smegmatis_MC2_155 TVLSRGRVIVDENSFHGQAG----------HGEYLKRSLSQYLI
MAB_2817c|M.abscessus_ATCC_199 TVLSRGKVIVDGDEYLGSKG----------DGVFLRRDLCQYLI
Mflv_3169|M.gilvum_PYR-GCK STWLRGT-KIDFTTPRGR---------------MLRRGGV----
. :