For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLAGAAEASPGVLTVDHAANTKFGRAMTTTTTGADIETSPPRFARAGVGAVAAVSSIALLVFLDRYGYFG DELYFLSAGRRLSFGYADQGPALPLIAHVLDSVAPGSLFVLRLPAVLATAAAIVISAQIAREFGGGRGAQ LLAAGGYATSPFLLVQGGQLATNSFDIPLWVLITWLLVRWARTRDDRLLLWAAVATAFALQIKWLVPILW ICVGLSVLLVGPRELLRRRFLWVGAAGAALTTIPMLVWQAGNGWPQLAMGSVIAAENDTVGGRPAFVPLA LFIAGLLGGVLLLCGVWALLRWEPLRPFRFVGVTVVLTFVVFMVLGGRAYYVAGVYAVAMAAGAVWLTRD GPRRRRKIVAVPLIAASAALAGWSLPWQPERDIPPVSADDGMATLTYGRFGWPELAEETAAAYRDLTDEE REWAVVIAESYWQASALDNYPDGDLPAVYSPGRGWGYFGAPPDDATTVLYVDRYPDTARELCAHVDPVGR ADARLGIPGVSRDVTIWKCVDPRQPWSTIWPSLRRL
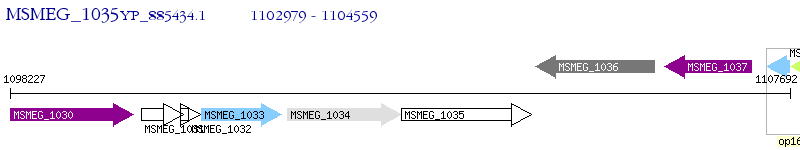
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1035 | - | - | 100% (526) | hypothetical protein MSMEG_1035 |
| M. smegmatis MC2 155 | MSMEG_2315 | - | 0.0 | 100.00% (526) | hypothetical protein MSMEG_2315 |
| M. smegmatis MC2 155 | MSMEG_3546 | - | 3e-81 | 40.47% (467) | hypothetical protein MSMEG_3546 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1277 | - | 6e-73 | 38.90% (455) | hypothetical protein MAB_1277 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1035|M.smegmatis_MC2_155 MLAGAAEASPGVLTVDHAANTKFGRAMTTTTTGADIETSPPR-FARAGVG
MAB_1277|M.abscessus_ATCC_1997 ----------------------MAAVTSRYDDGNNIPGVPPVCIARADLT
:. . : * :* ** :***.:
MSMEG_1035|M.smegmatis_MC2_155 AVAAVSSIALLVFLDRYGYFGDELYFLSAGR-RLSFGYADQGPALPLIAH
MAB_1277|M.abscessus_ATCC_1997 AITTLTFLVFGWASRGYGYFFDEAYFVVAGRDHLSWGYFDQPPLVPFIAG
*::::: :.: **** ** **: *** :**:** ** * :*:**
MSMEG_1035|M.smegmatis_MC2_155 VLDSVAPGSLFVLRLPAVLATAAAIVISAQIAREFGGGRGAQLLAAGGYA
MAB_1277|M.abscessus_ATCC_1997 VADNIAPGSLWVLRLAATAAVAIGTLMCGLIAAELGAGRLTQSLSAIAHA
* *.:*****:****.*. *.* . ::.. ** *:*.** :* *:* .:*
MSMEG_1035|M.smegmatis_MC2_155 TSPFLLVQGGQLATNSFDIPLWVLITWLLVRWARTR-----DDRLLLWAA
MAB_1277|M.abscessus_ATCC_1997 TAMFQILAH-WLQTPAIDPALWAVVCWLLVRWTRSLRERLPDDRLLLWAG
*: * :: * * ::* .**.:: ******:*: ********.
MSMEG_1035|M.smegmatis_MC2_155 VATAFALQIKWLVPILWICVGLSVLLVGPRELLRRRFLWVGAAGAALTTI
MAB_1277|M.abscessus_ATCC_1997 IVTAISMQTKFLIPALWFAVLLSAAVLGPRALLRRRQLWIGAAVVVVATA
:.**:::* *:*:* **:.* **. ::*** ***** **:*** ..::*
MSMEG_1035|M.smegmatis_MC2_155 PMLVWQAGNGWPQLAMGSVIAAENDTVGGRPAFVPLALFIAGLLGGVLLL
MAB_1277|M.abscessus_ATCC_1997 PTLIWQAAHGWPYLRMAAIVAAESERG---AALLGGMLVGSAVIGLVLLL
* *:***.:*** * *.:::***.: .*:: *. :.::* ****
MSMEG_1035|M.smegmatis_MC2_155 CGVWALLRWEPLRPFRFVGVTVVLTFVVFMVLGGRAYYVAGVYAVAMAAG
MAB_1277|M.abscessus_ATCC_1997 AGIVGLLASASLRPYRYLGSAAVLVGAFMFASHGRAHYAMGLYALLIGAG
.*: .** .***:*::* :.**. ..::. ***:*. *:**: :.**
MSMEG_1035|M.smegmatis_MC2_155 AVWLTRDGPRRRRKIVAVP-----LIAASAALAGWSLPWQPERDIP---P
MAB_1277|M.abscessus_ATCC_1997 AVTLSRWRPARPVARGVVYGFIGTVTVASVAMAVIRLPIVSEAAAARWFA
** *:* * * .* : .**.*:* ** .* . .
MSMEG_1035|M.smegmatis_MC2_155 VSADDGMATLTYGRFGWPELAEETAAAYRDLTDEEREWAVVIAESYWQAS
MAB_1277|M.abscessus_ATCC_1997 WAGGMGPTMLAEGDHSTEAFRQVARDTYDSLPADVRHRTALVVQIYPMAA
:.. * : *: * .. : : : :* .*. : *. :.::.: * *:
MSMEG_1035|M.smegmatis_MC2_155 ALDNYP-DGDLPAVYSPGRGWGYFGAPPDDATTVLYVDRYPDTARELCAH
MAB_1277|M.abscessus_ATCC_1997 AYDVEAGRAGISRAYSFHRGYYYFGAPPQSMTDMMYVG-VDDPDPKLAQG
* * . ..:. .** **: ******:. * ::**. *. :*.
MSMEG_1035|M.smegmatis_MC2_155 VDPVGRADARLGIPGVSRDVTIWKCVDPRQPWSTIWPSLRRL-
MAB_1277|M.abscessus_ATCC_1997 FRGVQRIELLHAAG--EDEAHVYRYYGRIAPWQQLWDDWRTYK
. * * : . . :. ::: . **. :* . *