For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAVMALQPVNRRSVPEDVFEQIVSEVLTGEMRPGESLPSERRLAEVLGVSRPAVREAIKRLTAAGLVEVR QGDATTVRDFRRHAGMDLLPRLLFRAGELDTSVVRSILETRLHNGPKVAELAAERRGAELVGQLTAVVHA LEAEIDPIQRQRHAMTFWDHVVDGADSIAFRLMFNTLRETYEPVLPALAAVMAGEVADPDAYRAITRAIS DRDADAARGAAQNLLEPATHALLGALDALEAAR
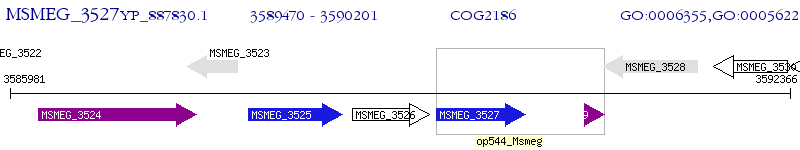
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3527 | - | - | 100% (243) | putative HTH-type transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2173 | - | 1e-14 | 28.63% (227) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2794 | - | 3e-14 | 30.36% (224) | transcriptional regulator, GntR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0601 | - | 1e-79 | 66.52% (233) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0016 | - | 4e-12 | 36.84% (133) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0586 | - | 1e-79 | 66.52% (233) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3018 | - | 1e-58 | 48.76% (242) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0820 | - | 7e-19 | 30.94% (223) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2542 | - | 9e-09 | 24.52% (208) | regulatory protein GntR, HTH |
| M. thermoresistible (build 8) | TH_1959 | - | 1e-97 | 75.00% (240) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (GNTR-FAMILY) |
| M. ulcerans Agy99 | MUL_4564 | - | 6e-19 | 30.94% (223) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2942 | - | 5e-96 | 73.75% (240) | regulatory protein GntR, HTH |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0601|M.bovis_AF2122/97 -------------MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSE
Rv0586|M.tuberculosis_H37Rv -------------MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSE
TH_1959|M.thermoresistible__bu -------------MALQPVNRRSVPEDVFDQIVGEVLSGQMQPGDALPSE
Mvan_2942|M.vanbaalenii_PYR-1 -------------MALRPVNRRSVPEDVFDQIISDVLSGEMRPGETLPSE
MSMEG_3527|M.smegmatis_MC2_155 ----------MAVMALQPVNRRSVPEDVFEQIVSEVLTGEMRPGESLPSE
MAB_3018|M.abscessus_ATCC_1997 -------------MALQPIARQSIPDEIFSQLAAQVLTGDRIPGDTLPSE
MMAR_0820|M.marinum_M ---------MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPE
MUL_4564|M.ulcerans_Agy99 ---------MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPE
Mflv_0016|M.gilvum_PYR-GCK -----MIGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNE
MAV_2542|M.avium_104 MPKREPLAPMATSQNNGAVRSPKTAEIVADTLRRMIVEGQLKDGDFLPYE
. : : : : *. :* *
Mb0601|M.bovis_AF2122/97 RRLAELLGVSRPAVREALKRLSAAGLVEVRQGDVTTVRDFRRHAGLDLLP
Rv0586|M.tuberculosis_H37Rv RRLAELLGVSRPAVREALKRLSAAGLVEVRQGDVTTVRDFRRHAGLDLLP
TH_1959|M.thermoresistible__bu RRLAEVLGVSRPAVREALKRLSAAGLVEVRQGEITTVRDYRRHAGLDLLP
Mvan_2942|M.vanbaalenii_PYR-1 RRLAEVLGVSRPAIREALKRLAAAGLIEVRQGDATTVRDFRRTAGLDLLP
MSMEG_3527|M.smegmatis_MC2_155 RRLAEVLGVSRPAVREAIKRLTAAGLVEVRQGDATTVRDFRRHAGMDLLP
MAB_3018|M.abscessus_ATCC_1997 RALAEALGVSRAAVREALGRLERSGLIQIRQGGSTVIRDYRSDAGFDVLP
MMAR_0820|M.marinum_M RELAEQLGVNRTSLRQGLARLQQMGLIETRQGSGNVVRDPQSLTHPAVVE
MUL_4564|M.ulcerans_Agy99 RELAEQLGVNRTSLRQGLARLQQMGLIETRQGSGNVVRDPQSLTHPAVVE
Mflv_0016|M.gilvum_PYR-GCK TVLVEKFGVGRNTVREAVRALAHAGILEVRQGDGTYVR-----ATSEVSG
MAV_2542|M.avium_104 AELMDHFQVSRPSLREAVRVLESDRLVEVRRGSRTGAR--VRVPGPEIVA
* : : *.* ::*:.: * ::: *:* . * . :
Mb0601|M.bovis_AF2122/97 RLLFRNGELDISVVRSILEARLRNFPKVAELAAER--NEPELAELLQDSL
Rv0586|M.tuberculosis_H37Rv RLLFRNGELDISVVRSILEARLRNFPKVAELAAER--NEPELAELLQDSL
TH_1959|M.thermoresistible__bu RLLFRAGELDVTVVRSILETRLHNGPKVAELAARR--RPPGLGALLDEAV
Mvan_2942|M.vanbaalenii_PYR-1 RLLIRDGDLDLAVVRSILETRLHNGPKVAELAAQR--RGPELSDELNTAM
MSMEG_3527|M.smegmatis_MC2_155 RLLFRAGELDTSVVRSILETRLHNGPKVAELAAER--RGAELVGQLTAVV
MAB_3018|M.abscessus_ATCC_1997 LLLAYGGDVDRRTLASVIEARAIIGPQVARLAAQRAHQTPGVADRLRAMT
MMAR_0820|M.marinum_M ALVRKLG---PDFLVEVLEIRAALGPLIGRLAAER--NSSEDAEALRAAL
MUL_4564|M.ulcerans_Agy99 ALVRKLG---PDFLVEVLEIRAALGPLIGRLAAER--NSSEDAEALRAAL
Mflv_0016|M.gilvum_PYR-GCK ALRRLCG----EELREVLEVRRALEVEAARLAASR--RTPADVAEMRGLL
MAV_2542|M.avium_104 RPAGFLLEMAGTTLADVMTARMGIEPYAARLLAED--GTAAAHRELRELI
: .:: * ..* * . :
Mb0601|M.bovis_AF2122/97 RALDTEEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAAL
Rv0586|M.tuberculosis_H37Rv RALDTEEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAAL
TH_1959|M.thermoresistible__bu QTLAAETDPVERQRHALRFWDHVVDGADSIAFRLMYNTLRATYEPALPAL
Mvan_2942|M.vanbaalenii_PYR-1 RALADEADPVAAQRHALDFWDHIVDGADSIAFRLMFNTLRATYEPALPAL
MSMEG_3527|M.smegmatis_MC2_155 HALEAEIDPIQRQRHAMTFWDHVVDGADSIAFRLMFNTLRETYEPVLPAL
MAB_3018|M.abscessus_ATCC_1997 SELEAEDDPLQRMWQALGFWELVIDTADSIAFRLVFNTMRDSYVRALDVL
MMAR_0820|M.marinum_M AEVADADTAPARQAADLAYFRVLIHGTRNRALGLLYRWVEQAFGGREHEL
MUL_4564|M.ulcerans_Agy99 AEVADADTAPARQAADLAYFRVLIHGTRNRALGLLYRWVEQAFGGREHDL
Mflv_0016|M.gilvum_PYR-GCK ARRDERDSAGDVEAFARIDTEFHLAVVRCARNNTLIELYRGLLEAVTASV
MAV_2542|M.avium_104 EEIPAAWETGKLGAASTALHRRLVELSGNATLTVIVGMLHEIFERHMTAA
. : : .
Mb0601|M.bovis_AF2122/97 TTTMTAAAKR------PSDYRKLADAICSGDPTGAKKAAQDLLELANTSL
Rv0586|M.tuberculosis_H37Rv TTTMTAAAKR------PSDYRKLADAICSGDPTGAKKAAQDLLELANTSL
TH_1959|M.thermoresistible__bu ATMMAAEVGR------PQAYRALAAAIEAGDPTAAEQSARDLLEPATAAL
Mvan_2942|M.vanbaalenii_PYR-1 ATMMAAEVGR------VDAYRAVAEAVLAGDPAAARDNAHRLLEPATTAL
MSMEG_3527|M.smegmatis_MC2_155 AAVMAGEVAD------PDAYRAITRAISDRDADAARGAAQNLLEPATHAL
MAB_3018|M.abscessus_ATCC_1997 VNVMAAEVGD------IGHYRALADAIALADPDAAQDAAAGMLALGTKAF
MMAR_0820|M.marinum_M TGAYEDADPV------IADLTAINDAVLAGDPETAAAAVEGYLNASALRM
MUL_4564|M.ulcerans_Agy99 TGAYEDADPV------IADLTAINDAVLAGDPETAAAAVEGYLNASALRM
Mflv_0016|M.gilvum_PYR-GCK AHTRHVPVD-------VCDHGVLLDHIAAGDPDLAARTAGEFLDRIINGL
MAV_2542|M.avium_104 FLSVQNVVPKAQYNKLMRSYTRLADLVAARDGAEAEAHWRRHMENASAEL
: : * * : :
Mb0601|M.bovis_AF2122/97 MAVLVSQASRQ----
Rv0586|M.tuberculosis_H37Rv MAVLVSQASRQ----
TH_1959|M.thermoresistible__bu LSALDELEAQK----
Mvan_2942|M.vanbaalenii_PYR-1 LDALHTLEEPR----
MSMEG_3527|M.smegmatis_MC2_155 LGALDALEAAR----
MAB_3018|M.abscessus_ATCC_1997 DKLLREMEKER----
MMAR_0820|M.marinum_M IMCYRAAYAGEDPER
MUL_4564|M.ulcerans_Agy99 IMCYRAAYAGEDPER
Mflv_0016|M.gilvum_PYR-GCK GSADSS---------
MAV_2542|M.avium_104 LKGHEKTKVRDIMH-