For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQISCAFATASDTPNHVALAESLGYRRAWLYDSPAVITDVWMTLARCAERTTRIGLGPGVLVPSLRHPMV NASAIAELATLAPGRVAVAVGTGFTGRLALGHRPIRWSDVADYVRCLKTLLAGETAEWEGARIRMLQLPG FGAPRPLRVPLLIAADGSRGRAVATELADGVFSVLPPRPTGPGVPNRRSQLVFGTVLSDGEDLASSRVVE TLASSAVLHYHRVYLLGGSAAVDELPGGRSWRKAVESYPADERHLAIHLVQPDRRHRPHLDSLIRPGMLV GTGAHVAKLIARYRAAGVTEIVYQPAGPDIERELHAFADAAGLTIGRRHTVCPLAST
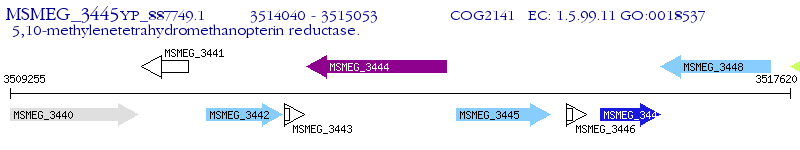
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3445 | - | - | 100% (337) | 5,10-methylenetetrahydromethanopterin reductase |
| M. smegmatis MC2 155 | MSMEG_2456 | - | 4e-14 | 25.15% (326) | 5,10-methylenetetrahydromethanopterin reductase |
| M. smegmatis MC2 155 | MSMEG_5520 | - | 5e-09 | 31.20% (125) | hypothetical protein MSMEG_5520 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0137c | fgd2 | 4e-11 | 29.76% (168) | putative f420-dependant glucose-6-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1539 | - | 1e-10 | 26.59% (346) | luciferase family protein |
| M. tuberculosis H37Rv | Rv0132c | fgd2 | 4e-11 | 29.76% (168) | putative f420-dependent glucose-6-phosphate dehydrogenase |
| M. leprae Br4923 | MLBr_00348 | - | 4e-09 | 25.08% (331) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4230c | - | 2e-10 | 32.45% (151) | F420-dependent glucose-6-phosphate dehydrogenase |
| M. marinum M | MMAR_0709 | fgd1 | 2e-09 | 31.88% (138) | F420-dependent glucose-6-phosphate dehydrogenase Fgd1 |
| M. avium 104 | MAV_5172 | - | 1e-11 | 26.46% (257) | F420-dependent glucose-6-phosphate dehydrogenase |
| M. thermoresistible (build 8) | TH_2264 | - | 3e-09 | 26.28% (331) | POSSIBLE COENZYME F420-DEPENDENT OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4782 | - | 4e-09 | 23.60% (356) | coenzyme F420-dependent oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_0698 | - | 1e-09 | 31.88% (138) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4230c|M.abscessus_ATCC_199 ------------------------------------------MARELKLG
Mvan_0698|M.vanbaalenii_PYR-1 ------------------------------------------MA-ELKLG
MMAR_0709|M.marinum_M ------------------------------------------MA-ELKLG
Mb0137c|M.bovis_AF2122/97 MTGISRRTFG-LAAGFGAIG----AGGLGGGCSTRSGPTPTPEPASRGVG
Rv0132c|M.tuberculosis_H37Rv MTGISRRTFG-LAAGFGAIG----AGGLGGGCSTRSGPTPTPEPASRGVG
MAV_5172|M.avium_104 MTGISRRTLGRLAVGAGVLASAVQACAKPGAGHGRSG--APPAPAGKGVG
Mflv_1539|M.gilvum_PYR-GCK ------------------------------------------MKLGLQLG
TH_2264|M.thermoresistible__bu --------------------------------------------------
MLBr_00348|M.leprae_Br4923 -----------------------------------MVERANAMKLGLQLG
MUL_4782|M.ulcerans_Agy99 ------------------------------------------MRTATTVE
MSMEG_3445|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4230c|M.abscessus_ATCC_199 YKASAEQFAPRELVELAVATESHGFDSATVSDHFQPWRYNGGHAPFSLAW
Mvan_0698|M.vanbaalenii_PYR-1 YKASAEQFAPRELVELAVAAEEHGMDSATVSDHFQPWRHEGGHAPFSLAW
MMAR_0709|M.marinum_M YKASAEQFAPRELVELAVAAEAHGMDSATVSDHFQPWRHEGGHAPFSLAW
Mb0137c|M.bovis_AF2122/97 VVLSHEQFRTDRLVAHAQAAEQAGFRYVWASDHLQPWQDNEGHSMFPWLT
Rv0132c|M.tuberculosis_H37Rv VVLSHEQFRTDRLVAHAQAAEQAGFRYVWASDHLQPWQDNEGHSMFPWLT
MAV_5172|M.avium_104 FVLSHEQFRTDRLVAHAQAAERAGFTHGWASDHIQPWQDNEGHSMFPWLT
Mflv_1539|M.gilvum_PYR-GCK YWGAQP---PENHAELVAAAEGAGFDTVFTAEAWG---------SDAYTP
TH_2264|M.thermoresistible__bu ----------------VAAAEEAGFDTVFTAEAWG---------SDAFTP
MLBr_00348|M.leprae_Br4923 YWAAQP---PGNAAELVAAAEGAGFDAVFTGEAWG---------SDAYTP
MUL_4782|M.ulcerans_Agy99 LSGGKD---GKDTVDFVVEAEKLGLHTCWVAEAWG---------SDAPSA
MSMEG_3445|M.smegmatis_MC2_155 MQISCAFATASDTPNHVALAESLGYRRAWLYDSPA-------VITDVWMT
. :* * :
MAB_4230c|M.abscessus_ATCC_199 MTAVGERTQRLQLGTSVLTPTFRYNPAVTAQAFATMGCLYPGRIFLGVGT
Mvan_0698|M.vanbaalenii_PYR-1 MTAVGERTKRLQLGTSVLTPTFRYNPAVIAQAFATMGCLYPGRIFLGVGT
MMAR_0709|M.marinum_M MTAVGERTKRMQLGTSVLTPTFRYNPAVIAQAFATMGCLYPNRIFLGVGT
Mb0137c|M.bovis_AF2122/97 LALVGNSTSSILFGTGVTCPIYRYHPATVAQAFASLAILNPGRVFLGLGT
Rv0132c|M.tuberculosis_H37Rv LALVGNSTSSILFGTGVTCPIYRYHPATVAQAFASLAILNPGRVFLGLGT
MAV_5172|M.avium_104 LALVGSSTSHVSFGTGVTCPTYRYHPATVAQAFASLAILNPGRVFLGVGT
Mflv_1539|M.gilvum_PYR-GCK LAWWGRETTRMRLGTSVIQLSAR-TPTACAMAALTLDHLSGGRHILGLGV
TH_2264|M.thermoresistible__bu LAWWGRQTTRMRLGTSVVQLSAR-TPTACAMASLTLDHLSGGRHILGLGV
MLBr_00348|M.leprae_Br4923 LAWWGSSTQRVRLGTSVVQLSAR-TPTACAMAALTLDHLSGGRHILGLGV
MUL_4782|M.ulcerans_Agy99 LGYIAARTERMQLGSGVLQVGTR-SPVLVAQTAITLSNLSCGRFLLGLGA
MSMEG_3445|M.smegmatis_MC2_155 LARCAERTTRIGLGPGVLVPSLR-HPMVNASAIAELATLAPGRVAVAVGT
: . * : :*..* * * * : : * .* :.:*.
MAB_4230c|M.abscessus_ATCC_199 GEALNEIATGYIGEWPEFKERFARLRESVRLMRELWT--------GERVD
Mvan_0698|M.vanbaalenii_PYR-1 GEALNEIATGYEGDWPEFKERYARLRESVRLMRDLWL--------GDRVD
MMAR_0709|M.marinum_M GEALNEVATGYQGVWPEFKERFARLRESVRLMRELWR--------GDRVD
Mb0137c|M.bovis_AF2122/97 GKRLNEQAATDT--FGNYRERHDRLIEAIVLIRQLWS--------GERIS
Rv0132c|M.tuberculosis_H37Rv GERLNEQAATDT--FGNYRERHDRLIEAIVLIRQLWS--------GERIS
MAV_5172|M.avium_104 GERLNEQATTNG--YGNYTERHDRLAEAIALIRQLWS--------GSRIS
Mflv_1539|M.gilvum_PYR-GCK SGPQ--VVEGWYG--AKFPKPLARTREYVDILRQVWAREAPVHSDGPHYP
TH_2264|M.thermoresistible__bu SGPQ--VVEGWYG--QRFPKPLARTREYIDIIRQVWAREAPVTSAGPHYP
MLBr_00348|M.leprae_Br4923 SGPQ--VVEGWYG--QPFPKPLSRTREYIDIVRQVWAREAPVVSDGQHYP
MUL_4782|M.ulcerans_Agy99 SGPR--VIEGLHG--VAFDRPLARLKETVEIVRQVFAGGKVTYS-GSEFQ
MSMEG_3445|M.smegmatis_MC2_155 GFTG-RLALGHR------PIRWSDVADYVRCLKTLLA--------GETAE
. : : :: : *
MAB_4230c|M.abscessus_ATCC_199 FEGEYYRTQGAAIYDVP-----EGGIPVYIAAGGAVVAKYAGRAGDGFIC
Mvan_0698|M.vanbaalenii_PYR-1 FEGEYYKTKGASIYDVP-----EGGIPIYIAAGGPQVAKYAGRAGDGFIC
MMAR_0709|M.marinum_M FDGEYYRLRGASIYDVP-----EGGVPVYIAAGGPAVAKYAGRAGDGFIC
Mb0137c|M.bovis_AF2122/97 FTGHYFRTDELKLYDTP-----AMPPPIFVAASGPQSATLAGRYGDGWIA
Rv0132c|M.tuberculosis_H37Rv FTGHYFRTDELKLYDTP-----AMPPPIFVAASGPQSATLAGRYGDGWIA
MAV_5172|M.avium_104 FSGRYFQTNSLKLYDVP-----ATPPPIFVAAGGPKSAKLAGQFGDGWIT
Mflv_1539|M.gilvum_PYR-GCK LPLTGEGTTGLGKNLKPITHPLRSDIPVMLGAEGPKNVALAAEICDGWLP
TH_2264|M.thermoresistible__bu LPLTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEIADGWLP
MLBr_00348|M.leprae_Br4923 LPLTGAGATGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLP
MUL_4782|M.ulcerans_Agy99 IPRRGGEARPMRLSTRP-----EYDIPIYLAALSPAMLRLTGRIADGWLG
MSMEG_3445|M.smegmatis_MC2_155 WEGARIRMLQLPGFGAPR----PLRVPLLIAADGSRGRAVATELADGVFS
* *: :.* .. : . ** :
MAB_4230c|M.abscessus_ATCC_199 TSGKGE---ELYSGKLIPAVREGAALAERSFDDIDRMIEIKISY--DPDP
Mvan_0698|M.vanbaalenii_PYR-1 TSGKGE---ELYKDKLIPAMREGAEAAGKNPDDIDRMIEIKISY--DTDP
MMAR_0709|M.marinum_M TSGKGE---ELYSEKLMPAVREGAAINNRNIDDIDKMIEIKISY--DPDP
Mb0137c|M.bovis_AF2122/97 QAR------DINDAKLLAAFAAGAQAAGRDPTTLGKRAELFAVV--GDD-
Rv0132c|M.tuberculosis_H37Rv QAR------DINDAKLLAAFAAGAQAAGRDPTTLGKRAELFAVV--GDD-
MAV_5172|M.avium_104 QSG------DVTNPKLLAAFGAGAQAAGRDVATLGKRAEMFAVV--GDD-
Mflv_1539|M.gilvum_PYR-GCK IFYSPR-IAGMYNEWLDEGFARPGARRTRETFEICATAQVVVTD---DRP
TH_2264|M.thermoresistible__bu IFYSPR-LADMYNEWLDEGFARPTARRSREDFEICATAQVVITD---DRA
MLBr_00348|M.leprae_Br4923 IFFSPR-MADMYNEWLDEGFARTGARRSREDFEICASAQIVVTE---DRA
MUL_4782|M.ulcerans_Agy99 TSFVPEGAGDAYFTHLDAGLAAAG--RSRADIDFCQGAEVAFASGVEELR
MSMEG_3445|M.smegmatis_MC2_155 VLPPRP--TGPGVPNRRSQLVFGTVLSDGEDLASSRVVETLASS------
. :
MAB_4230c|M.abscessus_ATCC_199 DKALENTRFWAPLSLTPEQKHSIHDPIEMERAADELPIEQVAKRWI----
Mvan_0698|M.vanbaalenii_PYR-1 ELALENTRFWAPLSLTAEQKHSIDDPIEMEKAADALPIEQVAKRWI----
MMAR_0709|M.marinum_M ELALNNTRFWAPLSLSAEQKRSIGDPIEMEAAAEALPIEQIAKRWI----
Mb0137c|M.bovis_AF2122/97 KAAARAADLWRFTAGAVDQP----NPVEIQRAAESNPIEKVLANWA----
Rv0132c|M.tuberculosis_H37Rv KAAARAADLWRFTAGAVDQP----NPVEIQRAAESNPIEKVLANWA----
MAV_5172|M.avium_104 AVAARAATLWRFTAGAVDQP----DPVDIQRAAESNPTDKVLGGWTGRHR
Mflv_1539|M.gilvum_PYR-GCK AIMELMKPHLALYMGGMGAEDTNFHADVYRRMGYAEVVDDVTRLFR----
TH_2264|M.thermoresistible__bu GTMEMIKPFLALYMGGMGAEEMNFHADVYRRMGYSEVVDDVTRLFR----
MLBr_00348|M.leprae_Br4923 AAFAGIKPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFR----
MUL_4782|M.ulcerans_Agy99 DMVAARKTELAFSLGGMGSASTNYYNQAYSRQGWSEVAAAVRERWQ----
MSMEG_3445|M.smegmatis_MC2_155 ---AVLHYHRVYLLGGSAAVDELPGGRSWRKAVESYPADERHLAIHLVQ-
MAB_4230c|M.abscessus_ATCC_199 -----------------------VASDPDEA-------------------
Mvan_0698|M.vanbaalenii_PYR-1 -----------------------VASDPDEA-------------------
MMAR_0709|M.marinum_M -----------------------VASDPDEA-------------------
Mb0137c|M.bovis_AF2122/97 -----------------------VGTDPGVH-------------------
Rv0132c|M.tuberculosis_H37Rv -----------------------VGTDPGVH-------------------
MAV_5172|M.avium_104 RGSARRGRAAGPRRGRRSLPALSAGRPDGRHRLLPGQCLTEIALNPLRAT
Mflv_1539|M.gilvum_PYR-GCK -----------------------SDRKDEAATVIP---------------
TH_2264|M.thermoresistible__bu -----------------------SGRKDEAAKVIP---------------
MLBr_00348|M.leprae_Br4923 -----------------------SNQKDKAAEIIP---------------
MUL_4782|M.ulcerans_Agy99 -----------------------RGDREGAARLVT---------------
MSMEG_3445|M.smegmatis_MC2_155 -----------------------PDRRHRPHL------------------
MAB_4230c|M.abscessus_ATCC_199 ------------VEKVADYVGWG-------LNHLVFHAPG--------HD
Mvan_0698|M.vanbaalenii_PYR-1 ------------VAKVKDYVDWG-------LNHLVFHAPG--------HD
MMAR_0709|M.marinum_M ------------VEKVGQYVAWG-------LNHLVLHAPG--------HD
Mb0137c|M.bovis_AF2122/97 ------------IGAVQAVLDAG--------AVPFLHFPQ--------DD
Rv0132c|M.tuberculosis_H37Rv ------------IGAVQAVLDAG--------AVPFLHFPQ--------DD
MAV_5172|M.avium_104 PVSPLPATNSRRIGQNRDVMDAVDPDSRHQLAVRMAELVRGMAAPRRLDQ
Mflv_1539|M.gilvum_PYR-GCK ------------DELVDDSAIVG------DLAYVKEQITAWE------AA
TH_2264|M.thermoresistible__bu ------------DELVDDAAIVG------DIDYVRKQIAAWE------AA
MLBr_00348|M.leprae_Br4923 ------------NELVDSTMIVG------DVDYVCKQIAAWS------AA
MUL_4782|M.ulcerans_Agy99 ------------DEMVLATTLIG------TEEMVRSRLVAWR------DA
MSMEG_3445|M.smegmatis_MC2_155 ------------DSLIRPGMLVG------TGAHVAKLIARYR------AA
MAB_4230c|M.abscessus_ATCC_199 QKRFLELFKTDLEP------------------------------------
Mvan_0698|M.vanbaalenii_PYR-1 QRRFLELFRRDLEP------------------------------------
MMAR_0709|M.marinum_M QLRFLELFEKDLAP------------------------------------
Mb0137c|M.bovis_AF2122/97 PITAIDFYRTNVLP------------------------------------
Rv0132c|M.tuberculosis_H37Rv PITAIDFYRTNVLP------------------------------------
MAV_5172|M.avium_104 VLAEVTAAAVEVIPGADIAGVLLVRKGGEFETLADTDSLAARLDVLQHDF
Mflv_1539|M.gilvum_PYR-GCK GVTMMVVGARSPEQ------------------------------------
TH_2264|M.thermoresistible__bu GVTMMVVGARSVEQ------------------------------------
MLBr_00348|M.leprae_Br4923 GVTMMMVSASSVEQ------------------------------------
MUL_4782|M.ulcerans_Agy99 GVNTVRLYPAGDTM------------------------------------
MSMEG_3445|M.smegmatis_MC2_155 GVTEIVYQPAGPDIER----------------------------------
:
MAB_4230c|M.abscessus_ATCC_199 ------------------RLRKLG--------------------------
Mvan_0698|M.vanbaalenii_PYR-1 ------------------RLRKLG--------------------------
MMAR_0709|M.marinum_M ------------------RLRRLG--------------------------
Mb0137c|M.bovis_AF2122/97 ------------------ELR-----------------------------
Rv0132c|M.tuberculosis_H37Rv ------------------ELR-----------------------------
MAV_5172|M.avium_104 GEGPCAQAALQETIVRSDDLRREPRWPCYAPAAVQLGVLSSLSFKLYTAD
Mflv_1539|M.gilvum_PYR-GCK ----------------IRDLADLV--------------------------
TH_2264|M.thermoresistible__bu ----------------IRDLAALI--------------------------
MLBr_00348|M.leprae_Br4923 ----------------VRDLAGLF--------------------------
MUL_4782|M.ulcerans_Agy99 ----------------DAKLSTLGRAIELVRAVR----------------
MSMEG_3445|M.smegmatis_MC2_155 ------------------ELHAFADAAGLTIGRRHTVCPLAST-------
*
MAB_4230c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0698|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0709|M.marinum_M --------------------------------------------------
Mb0137c|M.bovis_AF2122/97 --------------------------------------------------
Rv0132c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_5172|M.avium_104 RTAGALNLFSHRPDAWDTEAETIGSVFAAHAAAAILAGSRAEQLYSAVST
Mflv_1539|M.gilvum_PYR-GCK --------------------------------------------------
TH_2264|M.thermoresistible__bu --------------------------------------------------
MLBr_00348|M.leprae_Br4923 --------------------------------------------------
MUL_4782|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3445|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4230c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_0698|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0709|M.marinum_M --------------------------------------------------
Mb0137c|M.bovis_AF2122/97 --------------------------------------------------
Rv0132c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_5172|M.avium_104 RDRIGQAKGIIMERFGVDDVRAFDLLRRLSQESQVKLVEIAQQIIDTRGQ
Mflv_1539|M.gilvum_PYR-GCK --------------------------------------------------
TH_2264|M.thermoresistible__bu --------------------------------------------------
MLBr_00348|M.leprae_Br4923 --------------------------------------------------
MUL_4782|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_3445|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4230c|M.abscessus_ATCC_199 --
Mvan_0698|M.vanbaalenii_PYR-1 --
MMAR_0709|M.marinum_M --
Mb0137c|M.bovis_AF2122/97 --
Rv0132c|M.tuberculosis_H37Rv --
MAV_5172|M.avium_104 GA
Mflv_1539|M.gilvum_PYR-GCK --
TH_2264|M.thermoresistible__bu --
MLBr_00348|M.leprae_Br4923 --
MUL_4782|M.ulcerans_Agy99 --
MSMEG_3445|M.smegmatis_MC2_155 --